2FZV
 
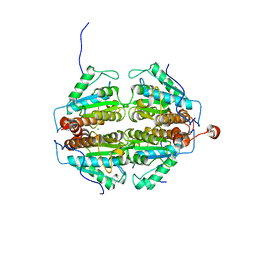 | | Crystal Structure of an apo form of a Flavin-binding Protein from Shigella flexneri | | 分子名称: | CALCIUM ION, CHLORIDE ION, putative arsenical resistance protein | | 著者 | Vorontsov, I.I, Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-02-10 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity
Protein Sci., 16, 2007
|
|
3CHH
 
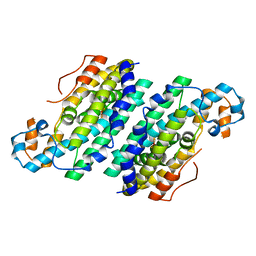 | | Crystal Structure of Di-iron AurF | | 分子名称: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | 著者 | Zhang, H, Brunzelle, J.S, Nair, S.K. | | 登録日 | 2008-03-09 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3KUU
 
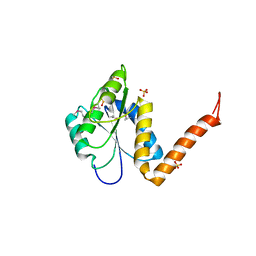 | | Structure of the PurE Phosphoribosylaminoimidazole Carboxylase Catalytic Subunit from Yersinia pestis | | 分子名称: | Phosphoribosylaminoimidazole carboxylase catalytic subunit PurE, SULFATE ION | | 著者 | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Kwon, K, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-11-27 | | 公開日 | 2009-12-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Structure of the PurE Phosphoribosylaminoimidazole Carboxylase Catalytic Subunit from Yersinia pestis
To be Published
|
|
4FAF
 
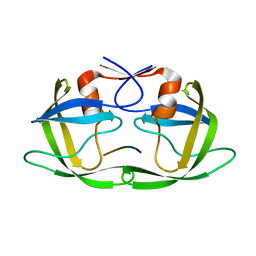 | | Substrate CA/p2 in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | 分子名称: | HIV-1 protease, substrate CA/p2 peptide | | 著者 | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | 登録日 | 2012-05-22 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
4EYR
 
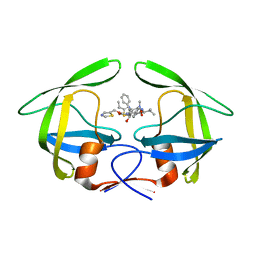 | | Crystal structure of multidrug-resistant clinical isolate 769 HIV-1 protease in complex with ritonavir | | 分子名称: | HIV-1 PROTEASE, RITONAVIR | | 著者 | Liu, Z, Yedidi, R.S, Wang, Y, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | 登録日 | 2012-05-01 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into the mechanism of drug resistance: X-ray structure analysis of multi-drug resistant HIV-1 protease ritonavir complex.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
4FAE
 
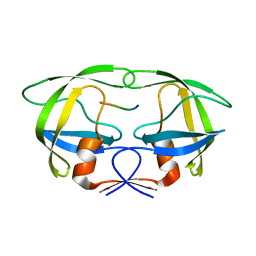 | | Substrate p2/NC in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | 分子名称: | HIV-1 protease, Substrate p2/NC peptide | | 著者 | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | 登録日 | 2012-05-22 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
2R31
 
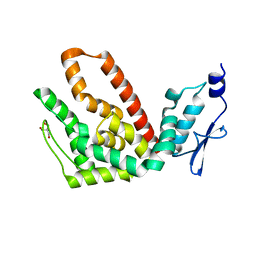 | | Crystal structure of atp12p from paracoccus denitrificans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP12 ATPase | | 著者 | Ludlam, A.V, Brunzelle, J.S, Gatti, D.L, Ackerman, S.H. | | 登録日 | 2007-08-28 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Chaperones of F1-ATPase.
J.Biol.Chem., 284, 2009
|
|
4MWA
 
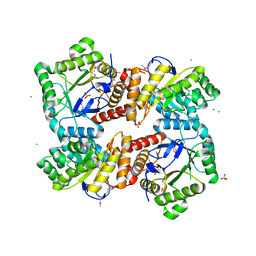 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | 分子名称: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | 著者 | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-24 | | 公開日 | 2013-10-09 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4N2Q
 
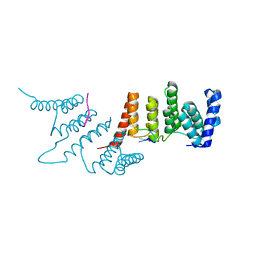 | | Crystal structure of THA8 in complex with Zm4 RNA | | 分子名称: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | 著者 | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | 登録日 | 2013-10-06 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4N2S
 
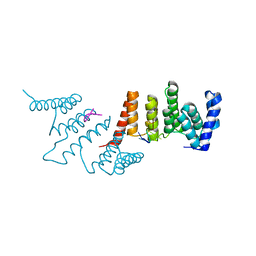 | | Crystal Structure of THA8 in complex with Zm1a-6 RNA | | 分子名称: | THA8 RNA binding protein, Zm1a-6 RNA | | 著者 | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | 登録日 | 2013-10-06 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6DK4
 
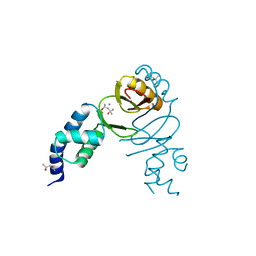 | | Crystal structure of Campylobacter jejuni peroxide stress regulator | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ferric uptake regulation protein, MANGANESE (II) ION, ... | | 著者 | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | 登録日 | 2018-05-28 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Crystal structure of Campylobacter jejuni peroxide regulator.
FEBS Lett., 592, 2018
|
|
6E2H
 
 | |
6E29
 
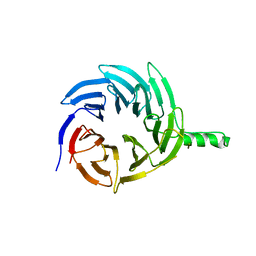 | |
8F4S
 
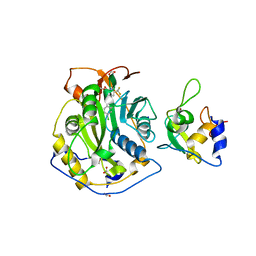 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | 分子名称: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-11-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | 分子名称: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-11-11 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8G28
 
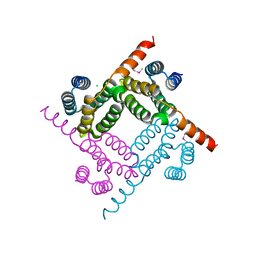 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | 分子名称: | ATPase, AAA family, CHLORIDE ION | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2023-02-03 | | 公開日 | 2023-02-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
8G22
 
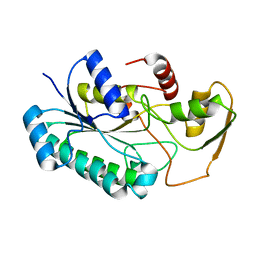 | | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae. | | 分子名称: | dTDP-4-dehydrorhamnose reductase | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2023-02-03 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae.
To Be Published
|
|
2BAS
 
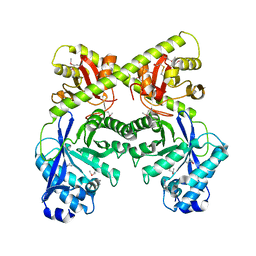 | | Crystal Structure of the Bacillus subtilis YkuI Protein, with an EAL Domain. | | 分子名称: | BETA-MERCAPTOETHANOL, YkuI protein | | 著者 | Minasov, G, Brunzelle, J.S, Shuvalova, L, Miller, D.J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-10-14 | | 公開日 | 2005-11-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains.
J.Biol.Chem., 284, 2009
|
|
7R6S
 
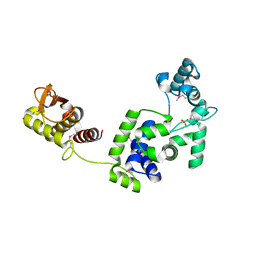 | | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia | | 分子名称: | Putative bacteriophage protein, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Brunzelle, J.S, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2021-06-23 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-02-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia
To Be Published
|
|
7S3G
 
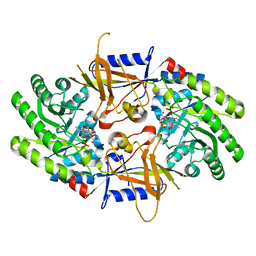 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | 分子名称: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | 登録日 | 2021-09-06 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3F
 
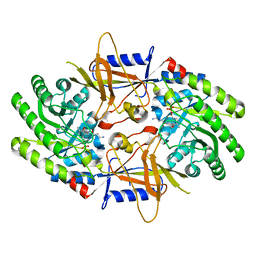 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | 分子名称: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | 登録日 | 2021-09-06 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7LJX
 
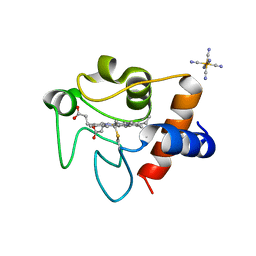 | | Oxidized rat cytochrome c mutant (K53Q) | | 分子名称: | Cytochrome c, somatic, HEME C, ... | | 著者 | Huttemann, M, Edwards, B.F.P, Brunzelle, J.S, Vaishnav, A. | | 登録日 | 2021-02-01 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Lysine 53 Acetylation of Cytochrome c in Prostate Cancer: Warburg Metabolism and Evasion of Apoptosis.
Cells, 10, 2021
|
|
7M74
 
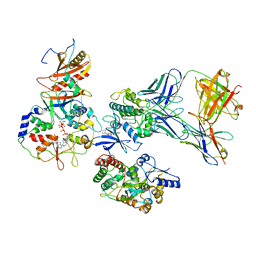 | | ATP-bound AMP-activated protein kinase | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | 登録日 | 2021-03-26 | | 公開日 | 2021-12-15 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
4RRU
 
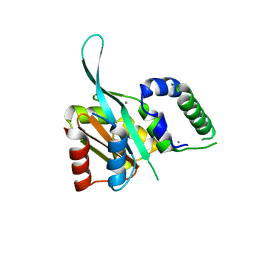 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | 分子名称: | CALCIUM ION, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-06 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RS9
 
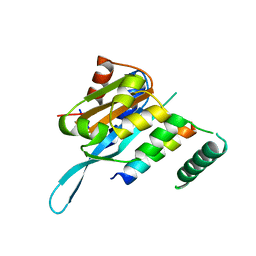 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | 分子名称: | Protein TIFY 7, Transcription factor MYC3 | | 著者 | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | 登録日 | 2014-11-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
