6TMW
 
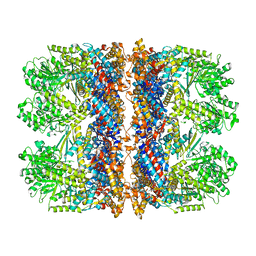 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6TMT
 
 | | Crystal structure of the chaperonin gp146 from the bacteriophage EL 2 (Pseudomonas aeruginosa) in presence of ATP-BeFx, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Paul, S.S, Wang, H, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6TMV
 
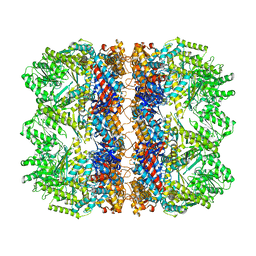 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in the apo state | | Descriptor: | Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
3O5I
 
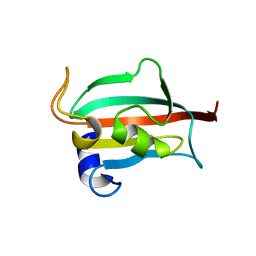 | | Fk1 domain of FKBP51, crystal form II | | Descriptor: | ACETATE ION, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5R
 
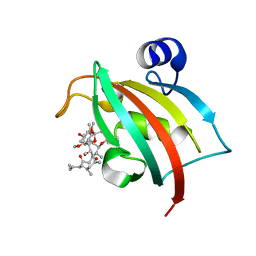 | | Complex of Fk506 with the Fk1 domain mutant A19T of FKBP51 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5F
 
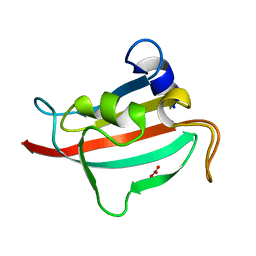 | | Fk1 domain of FKBP51, crystal form VII | | Descriptor: | CARBONATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5M
 
 | | Fk1 domain mutant A19T of FKBP51, crystal form II | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5D
 
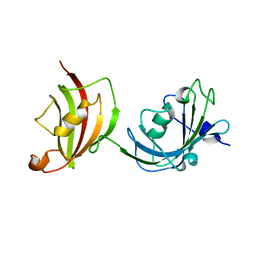 | | Crystal structure of a fragment of FKBP51 comprising the Fk1 and Fk2 domains | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O5K
 
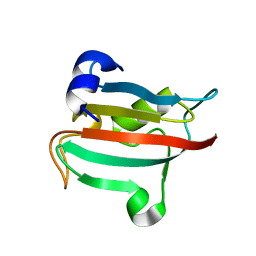 | | Fk1 domain of FKBP51, crystal form VIII | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4LAX
 
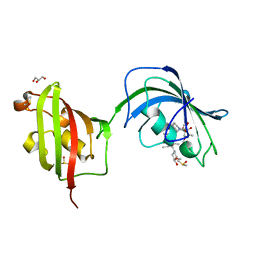 | | Crystal Structure Analysis of FKBP52, Complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4LAV
 
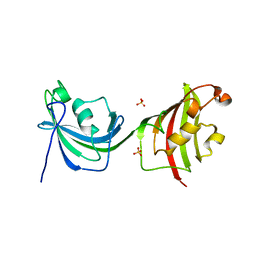 | | Crystal Structure Analysis of FKBP52, Crystal Form II | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, SULFATE ION | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4LAW
 
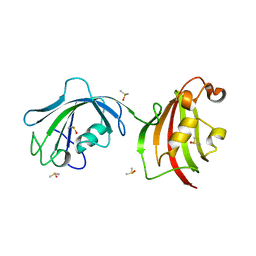 | | Crystal Structure Analysis of FKBP52, Crystal Form III | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4LAY
 
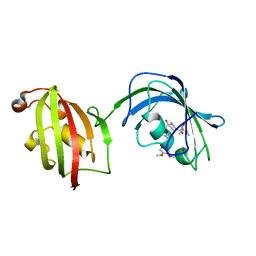 | | Crystal Structure Analysis of FKBP52, Complex with I63 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4W9O
 
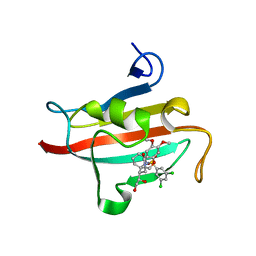 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4V94
 
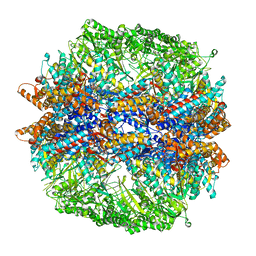 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
1KJT
 
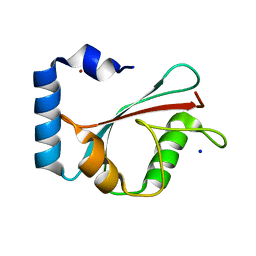 | | Crystal Structure of the GABA(A) Receptor Associated Protein, GABARAP | | Descriptor: | GABARAP, NICKEL (II) ION, SODIUM ION | | Authors: | Bavro, V.N, Sola, M, Bracher, A, Kneussel, M, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-12-05 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the GABA(A)-receptor-associated protein, GABARAP.
EMBO Rep., 3, 2002
|
|
9FVD
 
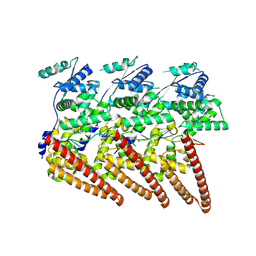 | | Cryo-EM structure of single-layered nucleoprotein-RNA helical assembly from Marburg virus, trimeric repeat unit | | Descriptor: | Nucleoprotein, RNA (5'-R(P*AP*GP*AP*CP*AP*CP*AP*CP*AP*AP*AP*AP*AP*CP*AP*AP*GP*A)-3') | | Authors: | Zinzula, L, Beck, F, Camasta, M, Bohn, S, Liu, C, Morado, D, Bracher, A, Plitzko, J.M, Baumeister, W. | | Deposit date: | 2024-06-26 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of single-layered nucleoprotein-RNA complex from Marburg virus
Nat Commun, 2024
|
|
4DRO
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH (1R)-3-(3,4-dimethoxyphenyl)-1-phenylpropyl (2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-{[(1R,2S)-2-ethyl-1-hydroxycyclohexyl](oxo)acetyl}piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
4DRH
 
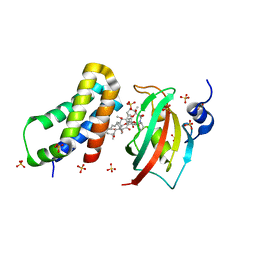 | | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR at low pH | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, SULFATE ION, ... | | Authors: | Maerz, A.M, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Large FK506-Binding Proteins Shape the Pharmacology of Rapamycin.
Mol.Cell.Biol., 33, 2013
|
|
6EKB
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
6EKC
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana bound to the octameric assembly of RbcL from Thermosynechococcus elongatus | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, Ribulose bisphosphate carboxylase large chain, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
7ZET
 
 | | Crystal structure of human Clusterin, crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin, ... | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Clusterin, crystal form I
To be published
|
|
7ZEU
 
 | |
4W9P
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
