6FM9
 
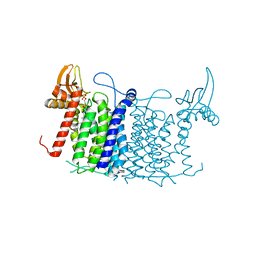 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
4RCI
 
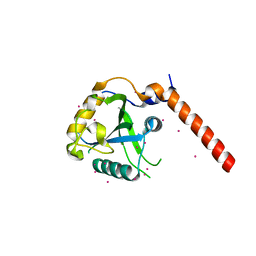 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
4QUU
 
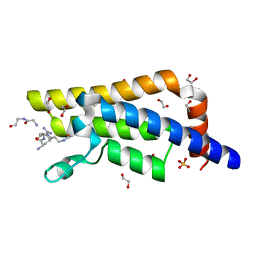 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) complexed with Histone H4-K(ac)5 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, Histone H4, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-12 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells.
J Mol Cell Biol, 8, 2016
|
|
3T7M
 
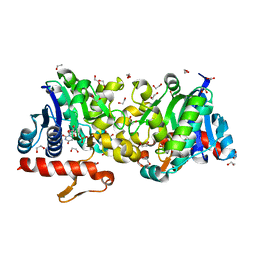 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a triclinic closed form | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4R3I
 
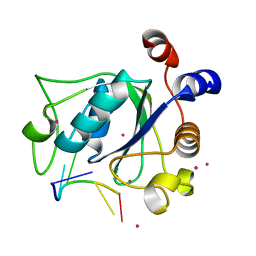 | | The crystal structure of an RNA complex | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Tempel, W, Xu, C, Liu, K, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
5O1O
 
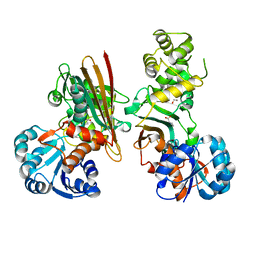 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Mathea, S, Velupillai, S, Strain-Damerell, C, Goubin, S, Kupinska, K, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound.
To Be Published
|
|
3HMF
 
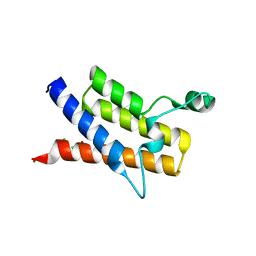 | | Crystal Structure of the second Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein polybromo-1, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
4NRA
 
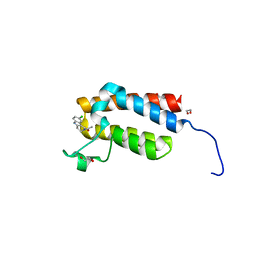 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-6 E11322 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-chloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Ferguson, F.M, Filippakopoulos, P, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
6CD8
 
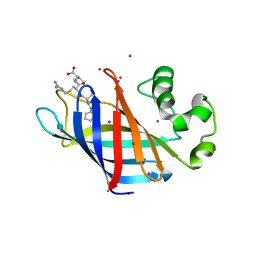 | | Complex of GID4 fragment with short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRV, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
3Q4U
 
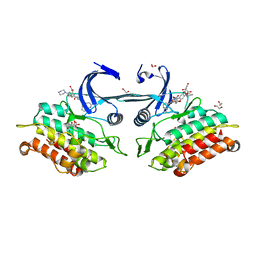 | | Crystal structure of the ACVR1 kinase domain in complex with LDN-193189 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C.D.O, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C.K, Krojer, T, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-24 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
3GEY
 
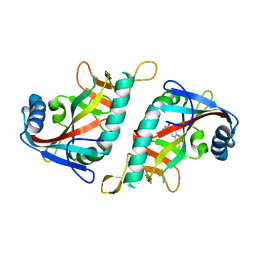 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment in complex with an inhibitor Pj34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Poly [ADP-ribose] polymerase 15 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
6SZM
 
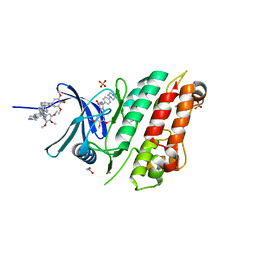 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2009 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-methyl-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl]piperazine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
3OP5
 
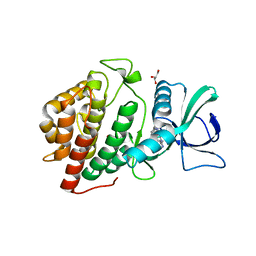 | | Human vaccinia-related kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase VRK1, ... | | Authors: | Allerston, C.K, Uttarkar, S, Savitsky, P, Elkins, J.M, Filippakopoulos, P, Krojer, T, Rellos, P, Fedorov, O, Eswaran, J, Brenner, B, Keates, T, Das, S, King, O, Chalk, R, Berridge, G, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
5SLK
 
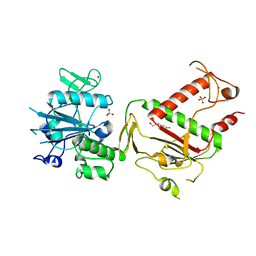 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1354370680 | | Descriptor: | 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLG
 
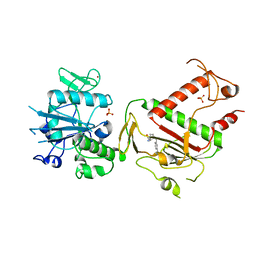 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z32400357 | | Descriptor: | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMC
 
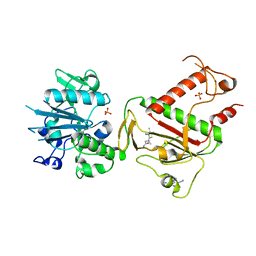 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2033637875 | | Descriptor: | N~2~-(4-cyano-3-methyl-1,2-thiazol-5-yl)-N~2~-methylglycinamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6BW3
 
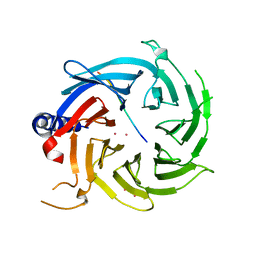 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
5SLF
 
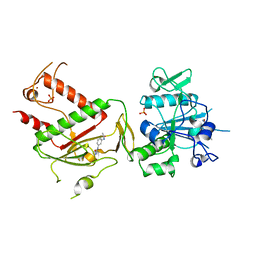 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z198195770 | | Descriptor: | N-[3-(carbamoylamino)phenyl]acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM5
 
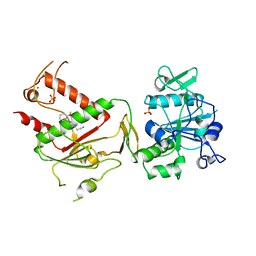 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434807 | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]ethanenitrile, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM8
 
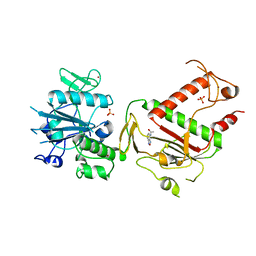 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2027158783 | | Descriptor: | N-(2,1,3-benzoxadiazol-4-yl)acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SME
 
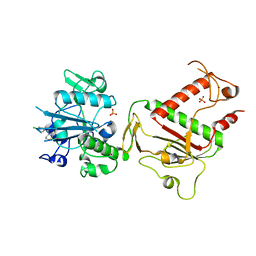 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z437584380 | | Descriptor: | (4-chlorophenyl)(thiomorpholin-4-yl)methanone, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLJ
 
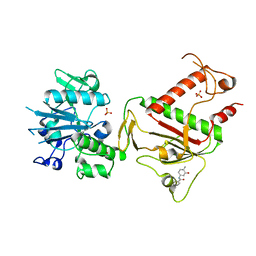 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1430613393 | | Descriptor: | 3-fluoro-N-(3-hydroxy-4-methylphenyl)benzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5O1P
 
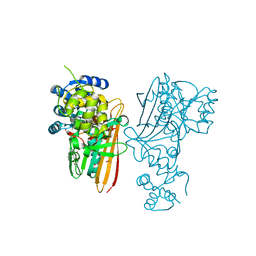 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
5SLI
 
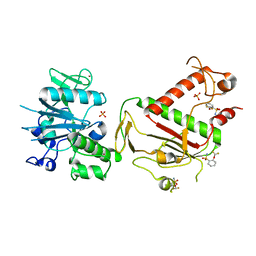 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1003146540 | | Descriptor: | 2-(difluoromethoxy)benzene-1-sulfonamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z71580604 | | Descriptor: | (2S)-N-(5-methylpyridin-2-yl)oxolane-2-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
