8CG9
 
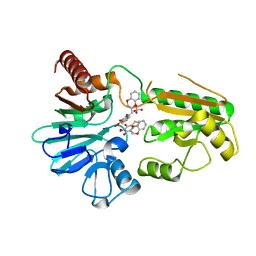 | | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, 1-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanyl-thieno[3,2-d]pyrimidine-2,4-dione, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
6WZZ
 
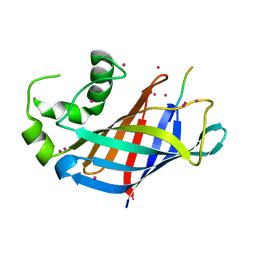 | | GID4 in complex with VGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZL2
 
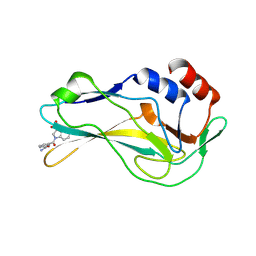 | | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-039-246-810 | | Descriptor: | (3S)-1-(3-aminocarbonylphenyl)carbonyl-2,3-dihydroindole-3-carboxylic acid, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
7ZK2
 
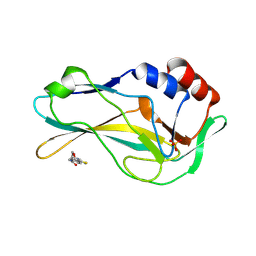 | | Crystal Structure of human Brachyury G177D variant in complex with CSC027898502 | | Descriptor: | N-[4-(2-morpholin-4-yl-1,3-thiazol-4-yl)phenyl]ethanamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
6I5H
 
 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5E1B
 
 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
4D0N
 
 | | AKAP13 (AKAP-Lbc) RhoGEF domain in complex with RhoA | | Descriptor: | 1,2-ETHANEDIOL, A-KINASE ANCHOR PROTEIN 13, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Abdul Azeez, K.R, Shrestha, L, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Klussmann, E, Elkins, J.M. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Rhoa : Akap-Lbc Dh-Ph Domain Complex.
Biochem.J., 464, 2014
|
|
5SMC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2033637875 | | Descriptor: | N~2~-(4-cyano-3-methyl-1,2-thiazol-5-yl)-N~2~-methylglycinamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z198195770 | | Descriptor: | N-[3-(carbamoylamino)phenyl]acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434807 | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]ethanenitrile, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLK
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1354370680 | | Descriptor: | 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2027158783 | | Descriptor: | N-(2,1,3-benzoxadiazol-4-yl)acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z32400357 | | Descriptor: | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1430613393 | | Descriptor: | 3-fluoro-N-(3-hydroxy-4-methylphenyl)benzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SME
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z437584380 | | Descriptor: | (4-chlorophenyl)(thiomorpholin-4-yl)methanone, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMK
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 NSP14 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5SMI
 
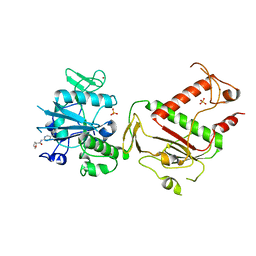 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z71580604 | | Descriptor: | (2S)-N-(5-methylpyridin-2-yl)oxolane-2-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLI
 
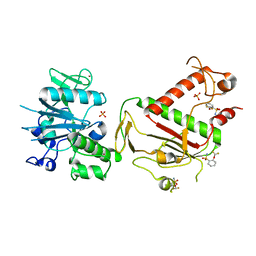 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1003146540 | | Descriptor: | 2-(difluoromethoxy)benzene-1-sulfonamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5DF6
 
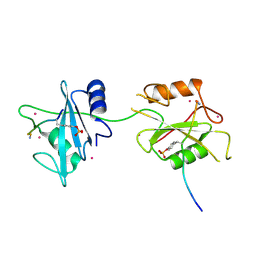 | | Crystal structure of PTPN11 tandem SH2 domains in complex with a TXNIP peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, UNKNOWN ATOM OR ION, txnip | | Authors: | Dong, A, Li, W, Tempel, W, Liu, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
3CEK
 
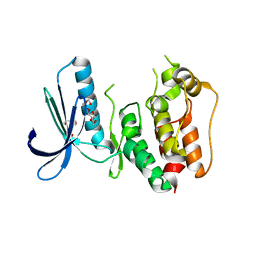 | | Crystal structure of human dual specificity protein kinase (TTK) | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, Roos, A, Pilka, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
5EQ0
 
 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
3MUJ
 
 | | Early B-cell factor 3 (EBF3) IPT/TIG and dimerization helices | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Transcription factor COE3 | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schueler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
6VIP
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008 | | Descriptor: | TP53-binding protein 1, UNKNOWN ATOM OR ION, {4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]phenyl}(4-ethylpiperazin-1-yl)methanone | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008
to be published
|
|
6NEY
 
 | | Crystal structure of TcBDF5, a bromodomain containing protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | Loppnau, P, Dong, A, Tempel, W, Lin, Y.H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-18 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of TcBDF5, a bromodomain containing protein from Trypanosoma cruzi
To Be Published
|
|
5OWQ
 
 | | Human STK10 bound to dovitinib | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Serine/threonine-protein kinase 10 | | Authors: | Szklarz, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2017-09-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human STK10 bound to dovitinib
To Be Published
|
|
