4LCT
 
 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6OT5
 
 | | Structure of the TRPV3 K169A sensitized mutant in the presence of 2-APB at 3.6 A resolution | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 3,Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
6OO4
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
5T7J
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
6OO3
 
 | | Cryo-EM structure of the C4-symmetric TRPV2/RTx complex in amphipol resolved to 2.9 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
5ACF
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5T7N
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6OT2
 
 | | Structure of the TRPV3 K169A sensitized mutant in apo form at 4.1 A resolution | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
7TQV
 
 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7TJ2
 
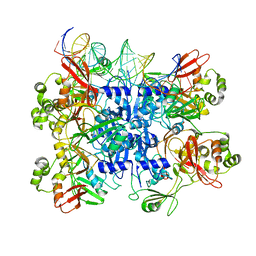 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (31-MER), Uridylate-specific endoribonuclease nsp15 | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
5ACH
 
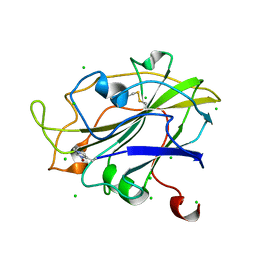 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACG
 
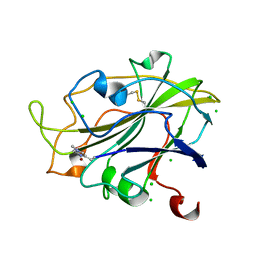 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACJ
 
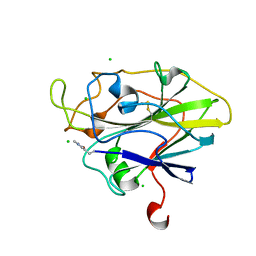 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
7T8C
 
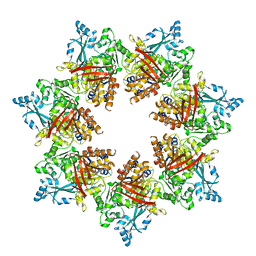 | | Heptameric Human Twinkle Helicase Clinical Variant W315L | | Descriptor: | Twinkle mtDNA helicase | | Authors: | Riccio, A.A, Bouvette, J, Krahn, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insight and characterization of human Twinkle helicase in mitochondrial disease.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T8B
 
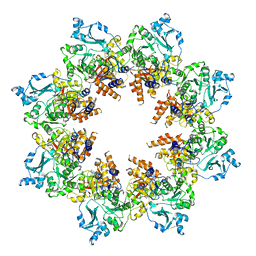 | | Octameric Human Twinkle Helicase Clinical Variant W315L | | Descriptor: | Twinkle mtDNA helicase | | Authors: | Riccio, A.A, Bouvette, J, Krahn, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight and characterization of human Twinkle helicase in mitochondrial disease.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2BHW
 
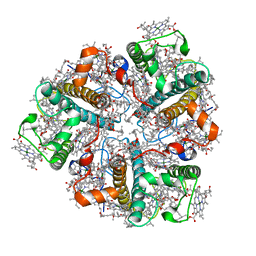 | | PEA LIGHT-HARVESTING COMPLEX II AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6'S,9R,9'R,13R,13'S)-4',5'-DIDEHYDRO-5',6',7',8',9,9',10,10',11,11',12,12',13,13',14,14',15,15'-OCTADECAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Standfuss, J, Terwisscha van Scheltinga, A.C, Lamborghini, M, Kuehlbrandt, W. | | Deposit date: | 2005-01-19 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of Photoprotection and Nonphotochemical Quenching in Pea Light-Harvesting Complex at 2.5 A Resolution.
Embo J., 24, 2005
|
|
7TPR
 
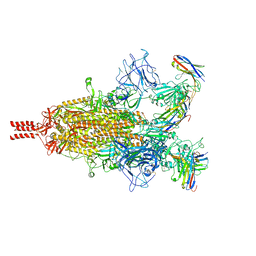 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
7TU6
 
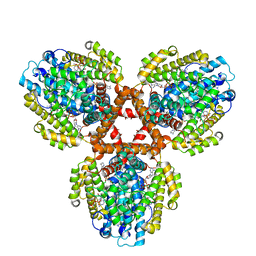 | | Structure of the L. blandensis dGTPase bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU7
 
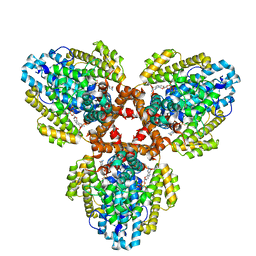 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU8
 
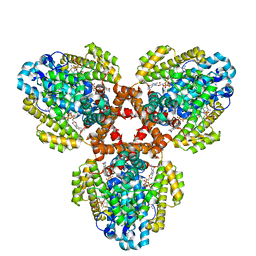 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP and dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU5
 
 | | Structure of the L. blandensis dGTPase in the apo form | | Descriptor: | MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7U65
 
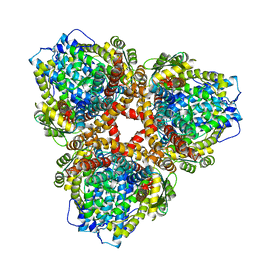 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, Inhibitor of dGTPase | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U67
 
 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
