1BW0
 
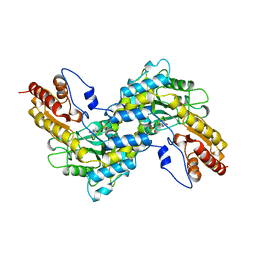 | | CRYSTAL STRUCTURE OF TYROSINE AMINOTRANSFERASE FROM TRYPANOSOMA CRUZI | | Descriptor: | PROTEIN (TYROSINE AMINOTRANSFERASE) | | Authors: | Blankenfeldt, W, Montemartini, M, Hunter, G.R, Kalisz, H.M, Nowicki, C, Hecht, H.J. | | Deposit date: | 1998-09-28 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi tyrosine aminotransferase: substrate specificity is influenced by cofactor binding mode.
Protein Sci., 8, 1999
|
|
1G1L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-GLUCOSE COMPLEX. | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CITRIC ACID, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naimsmith, J.H. | | Deposit date: | 2000-10-12 | | Release date: | 2000-12-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G23
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-16 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1FXO
 
 | |
1FZW
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). APO ENZYME. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1XUB
 
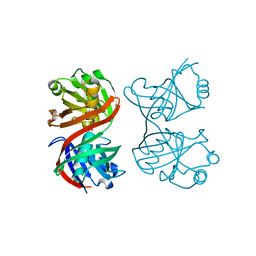 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2FXO
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FXM
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | MERCURY (II) ION, Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6NVX
 
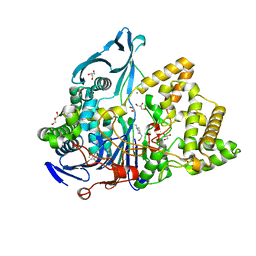 | | Crystal structure of penicillin G acylase from Bacillus sp. FJAT-27231 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
1G2V
 
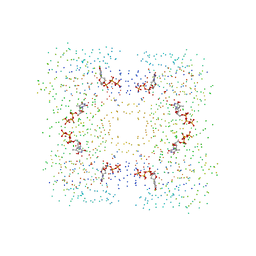 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
6NVW
 
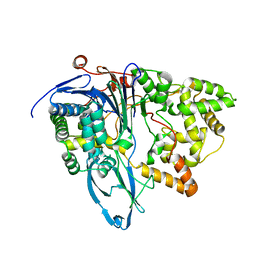 | | Crystal structure of penicillin G acylase from Bacillus megaterium | | Descriptor: | CALCIUM ION, Penicillin G acylase | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
6NVY
 
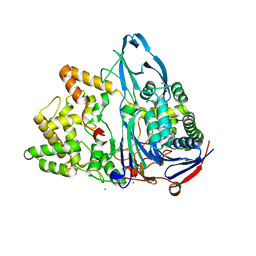 | | Crystal structure of penicillin G acylase from Bacillus thermotolerans | | Descriptor: | CALCIUM ION, GLYCEROL, Penicillin G acylase | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
1U1X
 
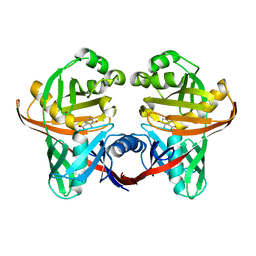 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1V
 
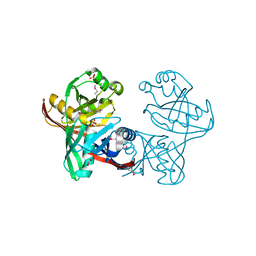 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1W
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1N2S
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-L-LYXO-4-HEXULOSE REDUCTASE (RMLD) IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Blankenfeldt, W, Kerr, I.D, Giraud, M.F, Mcmiken, H.J, Leonard, G.A, Whitfield, C, Messner, P, Graninger, M, Naismith, J.H. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variation on a Theme of SDR. dTDP-6-Deoxy-L- lyxo-4-Hexulose Reductase (RmlD) Shows a New Mg(2+)-Dependent Dimerization Mode
Structure, 10, 2002
|
|
1KBZ
 
 | | Crystal Structure of apo-dTDP-6-deoxy-L-lyxo-4-hexulose reductase (RmlD) from Salmonella enterica serovar Typhimurium | | Descriptor: | MAGNESIUM ION, dTDP-glucose oxidoreductase | | Authors: | Blankenfeldt, W, Kerr, I.D, Giraud, M.F, McMiken, H.J, Leonard, G.A, Whitfield, C, Messner, P, Graninger, M, Naismith, J.H. | | Deposit date: | 2001-11-07 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode.
Structure, 10, 2002
|
|
1KC3
 
 | | Crystal structure of dTDP-6-deoxy-L-lyxo-4-hexulose reductase (RmlD) in complex with NADPH and dTDP-L-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Blankenfeldt, W, Kerr, I.D, Giraud, M.F, McMiken, H.J, Leonard, G.A, Whitfield, C, Messner, P, Graninger, M, Naismith, J.H. | | Deposit date: | 2001-11-07 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode.
Structure, 10, 2002
|
|
1KC1
 
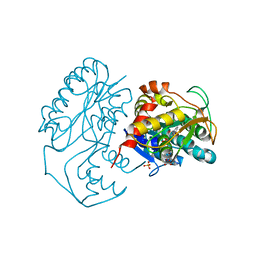 | | Crystal structure of dTDP-6-deoxy-L-lyxo-4-hexulose reductase (RmlD) in complex with NADPH | | Descriptor: | MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Kerr, I.D, Giraud, M.F, McMiken, H.J, Leonard, G.A, Whitfield, C, Messner, P, Graninger, M, Naismith, J.H. | | Deposit date: | 2001-11-07 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode.
Structure, 10, 2002
|
|
3SK2
 
 | |
3SK1
 
 | |
6ZTI
 
 | |
