3N6I
 
 | |
3N8R
 
 | |
3N7W
 
 | |
3N8L
 
 | |
2CNM
 
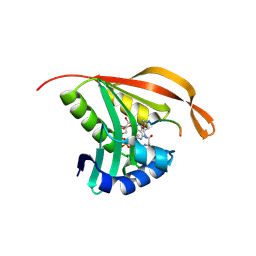 | | RimI - Ribosomal S18 N-alpha-protein acetyltransferase in complex with a bisubstrate inhibitor (Cterm-Arg-Arg-Phe-Tyr-Arg-Ala-N-alpha- acetyl-S-CoA). | | Descriptor: | 30S RIBOSOMAL PROTEIN S18, COENZYME A, MODIFICATION OF 30S RIBOSOMAL SUBUNIT PROTEIN S18 | | Authors: | Vetting, M.W, Yu, M, Bareich, D.C, Blanchard, J.S. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Rimi from Salmonella Typhimurium Lt2, the Gnat Responsible for N{Alpha}- Acetylation of Ribosomal Protein S18.
Protein Sci., 17, 2008
|
|
2C27
 
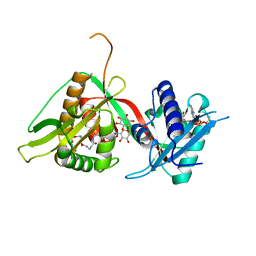 | | The Structure of Mycothiol Synthase in Complex with des- AcetylMycothiol and CoenzymeA. | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl 2-(L-cysteinylamino)-2-deoxy-alpha-D-glucopyranoside, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Vetting, M.W, Yu, M, Rendle, P.M, Blanchard, J.S. | | Deposit date: | 2005-09-26 | | Release date: | 2005-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Substrate-Induced Conformational Change of Mycobacterium Tuberculosis Mycothiol Synthase.
J.Biol.Chem., 281, 2006
|
|
4JLF
 
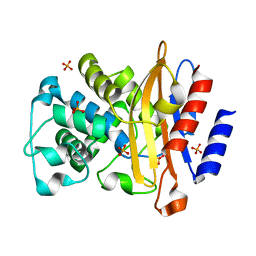 | | Inhibitor resistant (R220A) substitution in the Mycobacterium tuberculosis beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Kurz, S, Blanchard, J, Bonomo, R. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis beta-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of beta-lactam-beta-lactamase inhibitor combinations.
Antimicrob.Agents Chemother., 57, 2013
|
|
2JEV
 
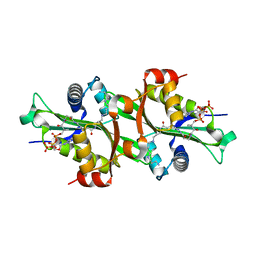 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
2GKJ
 
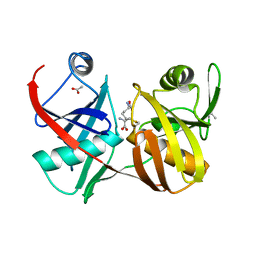 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor DL-AZIDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-02 | | Release date: | 2006-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1G2O
 
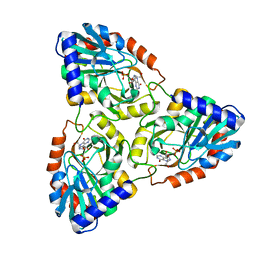 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
1I80
 
 | | CRYSTAL STRUCTURE OF M. TUBERCULOSIS PNP IN COMPLEX WITH IMINORIBITOL, 9-DEAZAHYPOXANTHINE AND PHOSPHATE ION | | Descriptor: | 9-DEAZAHYPOXANTHINE, IMINORIBITOL, PHOSPHATE ION, ... | | Authors: | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
2PR2
 
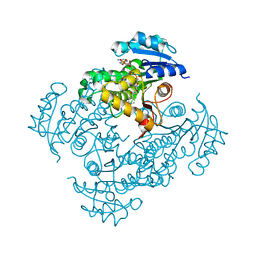 | |
2GKE
 
 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase, ... | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-01 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4HCX
 
 | |
1S7F
 
 | | RimL- Ribosomal L7/L12 alpha-N-protein acetyltransferase crystal form I (apo) | | Descriptor: | CHLORIDE ION, MALONIC ACID, acetyl transferase | | Authors: | Vetting, M.W, de Carvalho, L.P, Roderick, S.L, Blanchard, J.S. | | Deposit date: | 2004-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium.
J.Biol.Chem., 280, 2005
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
1ENZ
 
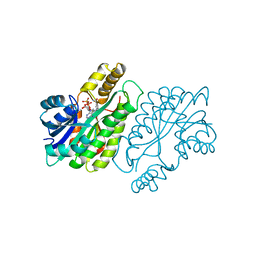 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
1ENY
 
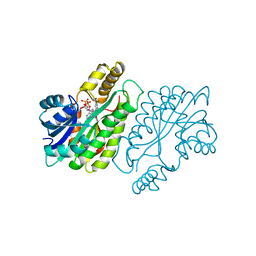 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
2Q9H
 
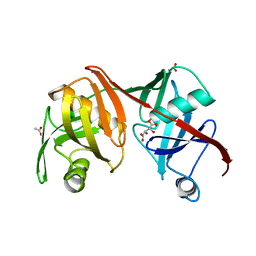 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2XTY
 
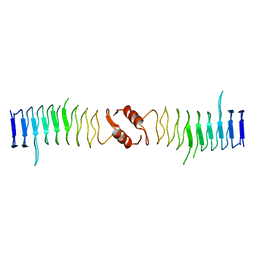 | | Structure of QnrB1 (R167E-Trypsin Treated), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1 | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-13 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
2XTW
 
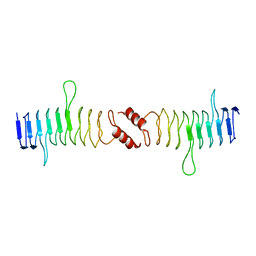 | | Structure of QnrB1 (Full length), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1 | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
2XTX
 
 | | Structure of QnrB1 (M102R-Trypsin Treated), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
1F06
 
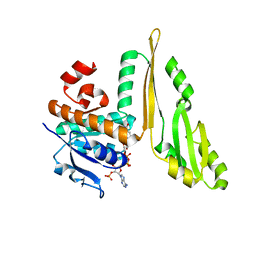 | | THREE DIMENSIONAL STRUCTURE OF THE TERNARY COMPLEX OF CORYNEBACTERIUM GLUTAMICUM DIAMINOPIMELATE DEHYDROGENASE NADPH-L-2-AMINO-6-METHYLENE-PIMELATE | | Descriptor: | L-2-AMINO-6-METHYLENE-PIMELIC ACID, MESO-DIAMINOPIMELATE D-DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cirilli, M, Scapin, G, Sutherland, A, Caplan, J.F, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 2000-05-14 | | Release date: | 2001-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-L-2-amino-6-methylene-pimelate.
Protein Sci., 9, 2000
|
|
2VZY
 
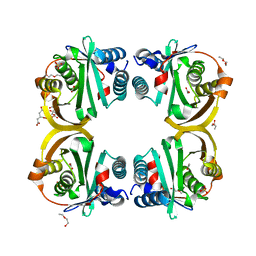 | | Crystal structure of Rv0802c from Mycobacterium tuberculosis in an unliganded form. | | Descriptor: | 2-ETHOXYETHANOL, ACETATE ION, CITRATE ANION, ... | | Authors: | Vetting, M.W, Errey, J.C, Blanchard, J.S. | | Deposit date: | 2008-08-07 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rv0802C from Mycobacterium Tuberculosis: The First Structure of a Succinyltransferase with the Gnat Fold.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VZZ
 
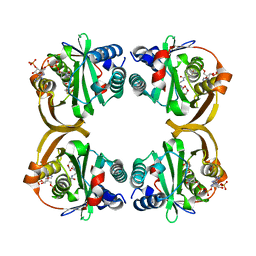 | |
