1V0C
 
 | | Structure of AAC(6')-Ib in complex with Kanamycin C and AcetylCoA. | | Descriptor: | AAC(6')-IB, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
3N8L
 
 | |
3N7W
 
 | |
3NC8
 
 | |
3NY4
 
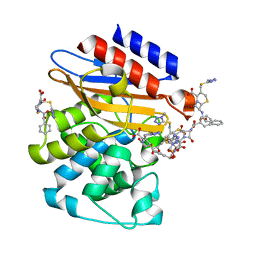 | | Crystal Structure of BlaC-K73A bound with Cefamandole | | Descriptor: | (6R,7R)-7-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-3-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
1P0H
 
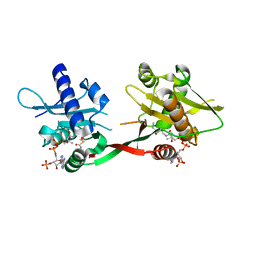 | | Crystal Structure of Rv0819 from Mycobacterium Tuberculosis MshD-Mycothiol Synthase Coenzyme A Complex | | Descriptor: | ACETYL COENZYME *A, COENZYME A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
1P9L
 
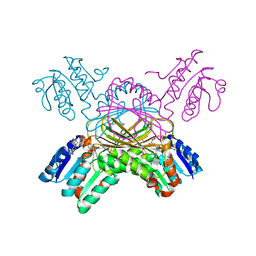 | | Structure of M. tuberculosis dihydrodipicolinate reductase in complex with NADH and 2,6 PDC | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PYRIDINE-2,6-DICARBOXYLIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes. Structural and mutagenic analysis of relaxed nucleotide specificity.
Biochemistry, 42, 2003
|
|
1OZP
 
 | | Crystal Structure of Rv0819 from Mycobacterium tuberculosis MshD-Mycothiol Synthase Acetyl-Coenzyme A Complex. | | Descriptor: | ACETYL COENZYME *A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
2AMB
 
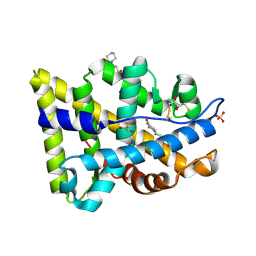 | | Crystal structure of human androgen receptor ligand binding domain in complex with tetrahydrogestrinone | | Descriptor: | 17-HYDROXY-18A-HOMO-19-NOR-17ALPHA-PREGNA-4,9,11-TRIEN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
2VQY
 
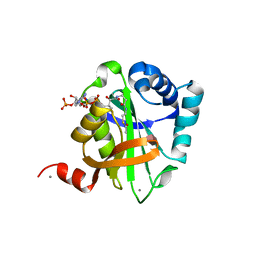 | | Structure of AAC(6')-Ib in complex with Parmomycin and AcetylCoA. | | Descriptor: | AAC(6')-IB, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Vetting, M.W, Park, C.H, Hedge, S.S, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic and Structural Analysis of Aminoglycoside N-Acetyltransferase Aac(6')-Ib and its Bifunctional, Fluoroquinolone-Active Aac(6')-Ib-Cr Variant.
Biochemistry, 47, 2008
|
|
2VBQ
 
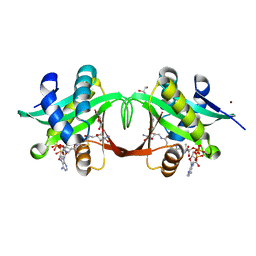 | | Structure of AAC(6')-Iy in complex with bisubstrate analog CoA-S- monomethyl-acetylneamine. | | Descriptor: | (3R,9Z)-17-[(2R,3S,4R,5R,6R)-5-amino-6-{[(1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl]oxy}-3,4-dihydroxytetrahydro-2H-pyran-2-yl]-3-hydroxy-2,2-dimethyl-4,8,15-trioxo-12-thia-5,9,16-triazaheptadec-9-en-1-yl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, AMINOGLYCOSIDE 6'-N-ACETYLTRANSFERASE, GLYCEROL, ... | | Authors: | Vetting, M.W, Magalhaes, M.L, Freiburger, L, Gao, F, Auclair, K, Blanchard, J.S. | | Deposit date: | 2007-09-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and Structural Analysis of Bisubstrate Inhibition of the Salmonella Enterica Aminoglycoside 6'-N-Acetyltransferase.
Biochemistry, 47, 2008
|
|
2W7Z
 
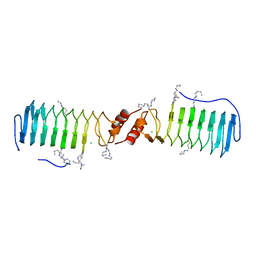 | | Structure of the pentapeptide repeat protein EfsQnr, a DNA gyrase inhibitor. Free amines modified by cyclic pentylation with glutaraldehyde. | | Descriptor: | CHLORIDE ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Vetting, M.W, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2009-01-06 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of a Pentapeptide-Repeat Protein by Reductive Cyclic Pentylation of Free Amines with Glutaraldehyde.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2X9Q
 
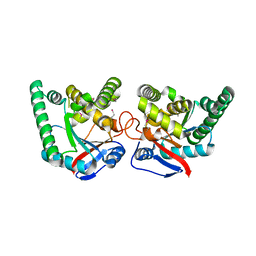 | |
2XT2
 
 | | Structure of the pentapeptide repeat protein AlbG, a resistance factor for the topoisomerase poison albicidin. | | Descriptor: | MCBG-LIKE PROTEIN, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2010-10-05 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Pentapeptide-Repeat Proteins that Act as Topoisomerase Poison Resistance Factors Have a Common Dimer Interface.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2J8I
 
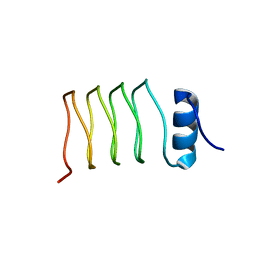 | | Structure of NP275, a pentapeptide repeat protein from Nostoc punctiforme | | Descriptor: | CHLORIDE ION, NP275 | | Authors: | Vetting, M.W, Hegde, S.S, Hazleton, K.Z, Blanchard, J.S. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Fusion of Two Pentapeptide Repeat Proteins, Np275 and Np276, from Nostoc Punctiforme: Resurrection of an Ancestral Protein.
Protein Sci., 16, 2007
|
|
2J8K
 
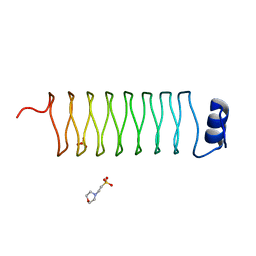 | | Structure of the fusion of NP275 and NP276, pentapeptide repeat proteins from Nostoc punctiforme | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NP275-NP276, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Hazleton, K.Z, Blanchard, J.S. | | Deposit date: | 2006-10-25 | | Release date: | 2006-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of the Fusion of Two Pentapeptide Repeat Proteins, Np275 and Np276, from Nostoc Punctiforme: Resurrection of an Ancestral Protein.
Protein Sci., 16, 2007
|
|
