5DK6
 
 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
4ZQ8
 
 | | Crystal structure of a terpene synthase from Streptomyces lydicus, target EFI-540129 | | Descriptor: | Isoprenoid Synthase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-08 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a terpene synthase from Streptomyces lydicus, target EFI-540129
To be published
|
|
4NHZ
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
4NSN
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry | | Descriptor: | ADENINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry.
To be Published
|
|
4NS1
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
To be Published
|
|
4WR2
 
 | | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Pyrimidine-specific ribonucleoside hydrolase RihA | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site
To be published
|
|
4WK5
 
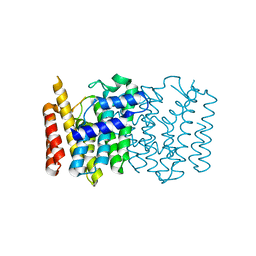 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
4FI4
 
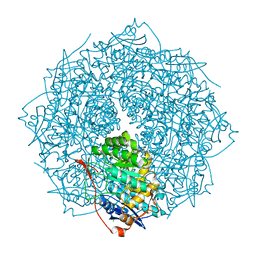 | | Crystal structure of mannonate dehydratase PRK15072 (TARGET EFI-502214) from Caulobacter sp. K31 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enolase Prk15072 (Target Efi-502214) from Caulobacter Sp. K31
To be Published
|
|
4GF0
 
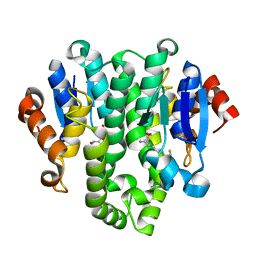 | | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione
To be Published
|
|
4G9H
 
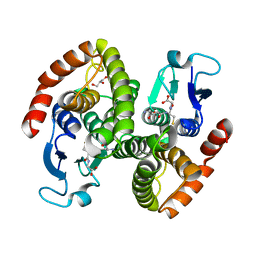 | | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target efi-501894, with bound glutathione
To be Published
|
|
3UAR
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501774) from methylococcus capsulatus str. bath with gsh bound | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Methylococcus Capsulatus
To be Published
|
|
3UBL
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans with gsh bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
3UIF
 
 | | CRYSTAL STRUCTURE OF putative sulfonate ABC transporter, periplasmic sulfonate-binding protein SsuA from Methylobacillus flagellatus KT | | Descriptor: | GLYCEROL, SULFATE ION, Sulfonate ABC transporter, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative sulfonate ABC transporter, periplasmic sulfonate-binding protein
SsuA from Methylobacillus flagellatus KT
To be Published
|
|
3UOG
 
 | | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Alcohol dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3UJ2
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE | | Descriptor: | Enolase 1, MAGNESIUM ION, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE
to be published
|
|
3UGV
 
 | | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG | | Descriptor: | CHLORIDE ION, Enolase, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an enolase from alpha pretobacterium bal199 (EFI TARGET EFI-501650) with bound MG
to be published
|
|
3EB3
 
 | | Voltage-dependent K+ channel beta subunit (W121A) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
3UF0
 
 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
3EAU
 
 | | Voltage-dependent K+ channel beta subunit in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
3EB4
 
 | | Voltage-dependent K+ channel beta subunit (I211R) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
4RK2
 
 | | Crystal structure of sugar transporter RHE_PF00321 from Rhizobium etli, target EFI-510806, an open conformation | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative sugar ABC transporter, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Sugar Transporter RHE_Pf00321 from Rhizobium Etli, Target EFI-510806
To be Published
|
|
4RKR
 
 | | Crystal structure of LacI family transcriptional regulator from Arthrobacter sp. FB24, target EFI-560007, complex with lactose | | Descriptor: | Transcriptional regulator, LacI family, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator PurR from Arthrobacter Sp, Target Nysgxrc 11027R
To be Published
|
|
4RXT
 
 | | Crystal structure of carbohydrate transporter solute binding protein Arad_9553 from Agrobacterium Radiobacter, Target EFI-511541, in complex with D-arabinose | | Descriptor: | Sugar ABC transporter, alpha-L-arabinopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Sugar Binding Transporter Arad_9553 from Agrobacterium Radiobacter, Target Efi-511541
To be Published
|
|
4RY8
 
 | | Crystal structure of 5-methylthioribose transporter solute binding protein TLET_1677 from Thermotoga lettingae TMO TARGET EFI-511109 in complex with 5-methylthioribose | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, Periplasmic binding protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of 5-methylthioribose binding protein TLET_1677 from Thermotoga lettingae TARGET EFI-511109
To be Published
|
|
4RK9
 
 | | CRYSTAL STRUCTURE OF SUGAR TRANSPORTER BL01359 FROM Bacillus licheniformis, TARGET EFI-510856, IN COMPLEX WITH STACHYOSE | | Descriptor: | Carbohydrate ABC transporter substrate-binding protein MsmE, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-[beta-D-fructofuranose-(2-1)]alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CRYSTAL STRUCTURE OF SUGAR TRANSPORTER BL01359 FROM Bacillus licheniformis, TARGET EFI-510856
To be Published
|
|
