2CTX
 
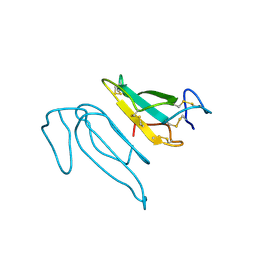 | | THE REFINED CRYSTAL STRUCTURE OF ALPHA-COBRATOXIN FROM NAJA NAJA SIAMENSIS AT 2.4-ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-COBRATOXIN | | Authors: | Betzel, C, Lange, G, Pal, G.P, Wilson, K.S, Maelicke, A, Saenger, W. | | Deposit date: | 1991-09-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The refined crystal structure of alpha-cobratoxin from Naja naja siamensis at 2.4-A resolution.
J.Biol.Chem., 266, 1991
|
|
353D
 
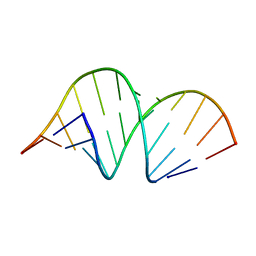 | | CRYSTAL STRUCTURE OF DOMAIN A OF THERMUS FLAVUS 5S RRNA AND THE CONTRIBUTION OF WATER MOLECULES TO ITS STRUCTURE | | Descriptor: | RNA (5'-R(*AP*UP*CP*CP*CP*CP*CP*GP*UP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*GP*CP*GP*GP*GP*GP*GP*AP*U)-3') | | Authors: | Betzel, C, Lorenz, S, Furste, J.P, Bald, R, Zhang, M, Schneider, T.R, Wilson, K.S, Erdmann, V.A. | | Deposit date: | 1997-09-29 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure.
FEBS Lett., 351, 1994
|
|
1IC6
 
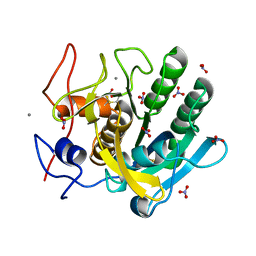 | | STRUCTURE OF A SERINE PROTEASE PROTEINASE K FROM TRITIRACHIUM ALBUM LIMBER AT 0.98 A RESOLUTION | | Descriptor: | CALCIUM ION, NITRATE ION, PROTEINASE K | | Authors: | Betzel, C, Gourinath, S, Kumar, P, Kaur, P, Perbandt, M, Eschenburg, S, Singh, T.P. | | Deposit date: | 2001-03-30 | | Release date: | 2001-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution.
Biochemistry, 40, 2001
|
|
2PRK
 
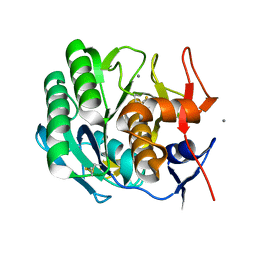 | |
1SVN
 
 | | SAVINASE | | Descriptor: | CALCIUM ION, SAVINASE (TM) | | Authors: | Betzel, C, Klupsch, S, Papendorf, G, Hastrup, S, Branner, S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the alkaline proteinase Savinase from Bacillus lentus at 1.4 A resolution.
J.Mol.Biol., 223, 1992
|
|
1PEK
 
 | | STRUCTURE OF THE COMPLEX OF PROTEINASE K WITH A SUBSTRATE-ANALOGUE HEXA-PEPTIDE INHIBITOR AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | D-DAL-ALA-NH2, PEPTIDE PRO-ALA-PRO-PHE, PROTEINASE K | | Authors: | Betzel, C, Singh, T.P, Visanji, M, Peters, K, Fittkau, S, Saenger, W, Wilson, K.S. | | Deposit date: | 1993-01-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution.
J.Biol.Chem., 268, 1993
|
|
4YEK
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3T62
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean Sea anemone Stichodactyla helianthus in complex with bovine chymotrypsin | | Descriptor: | Chymotrypsinogen A, Kunitz-type proteinase inhibitor SHPI-1, SULFATE ION | | Authors: | Garcia-Fernandez, R, Dominguez, R, Oberthuer, D, Pons, T, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into chymotrypsin inhibition by the Kunitz-type inhibitor-1 from the marine invertebrate Stichodactyla helianthus
To be Published
|
|
1ZL7
 
 | | Crystal structure of catalytically-active phospholipase A2 with bound calcium | | Descriptor: | CALCIUM ION, GLYCEROL, hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
2X2Q
 
 | | Crystal structure of an 'all locked' LNA duplex at 1.9 angstrom resolution | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), LOCKED NUCLEIC ACID DERIVED FROM TRNA SER ACCEPTOR STEM MICROHELIX, ... | | Authors: | Eichert, A, Behling, K, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2010-01-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of an 'All Locked' Nucleic Acid Duplex.
Nucleic Acids Res., 38, 2010
|
|
1QL0
 
 | |
4U2K
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with anticancer compound at 2.13 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-2,3-diaminopropyl]-5-fluoropyrimidine-2,4(1H,3H)-dione, 1-[(2S)-2,3-diaminopropyl]-5-fluoropyrimidine-2,4(1H,3H)-dione, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with new anticancer compound at 1.17 A resolution
To Be Published
|
|
6EYP
 
 | | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Prokofev, I.I, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A
To Be Published
|
|
6FJS
 
 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
4EB2
 
 | | Crystal structure Mistletoe Lectin I from Viscum album in complex with n-acetyl-d-glucosamine at 1.94 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Laskov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure Mistletoe Lectin I from Viscum album in complex with N-acetyl-D-glucosamine at 1.94 A resolution.
To be Published
|
|
5OLN
 
 | | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis at 1.88 A | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Pyrimidine-nucleoside phosphorylase, ... | | Authors: | Balaev, V.V, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of pyrimidine-nucleoside phosphorylase from Bacillus subtilis in complex with imidazole and sulfate.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8R7M
 
 | | CTX-M14 in complex with boric acid and 1,2-diol boric ester | | Descriptor: | BORIC ACID, Beta-lactamase, GLYCEROL, ... | | Authors: | Werner, N, Prester, A, Hinrichs, W, Perbandt, M, Betzel, C. | | Deposit date: | 2023-11-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
9KX8
 
 | | Mistletoe Lectin I from Viscum album complexed with epimer form of lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside-specific lectin 1 chain A isoform 1, ... | | Authors: | Saeed, A, Betzel, C, Brognaro, H, Rajaiah Prabhu, P, Alves Franca, B, Khaliq, B, Mehmood, S, Akrem, A. | | Deposit date: | 2024-12-06 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Mistletoe Lectin I from Viscum album complexed with epimer form of lactose
To Be Published
|
|
4E3Y
 
 | | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-ray structure of the Serratia marcescens endonuclease at 0.95 A resolution
To be Published
|
|
5Z4V
 
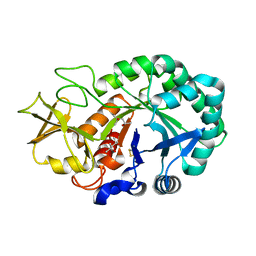 | | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Sharma, P, Singh, P.K, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40
To Be Published
|
|
9LH0
 
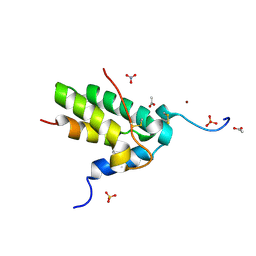 | | High-resolution crystal structure of an isoform of Chitin Binding Protein from Iberis umbellata L. | | Descriptor: | ACETIC ACID, Chitin Binding Protein, LITHIUM ION, ... | | Authors: | Saeed, A, Betzel, C, Brognaro, H, Rajaiah Prabhu, P, Alves Franca, B, Mehmood, S, Khaliq, B, Ishaq, U, Akrem, A. | | Deposit date: | 2025-01-11 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Crystal structure of Chitin Binding Protein from Iberis umbellata L.
To Be Published
|
|
5IN2
 
 | |
9GVH
 
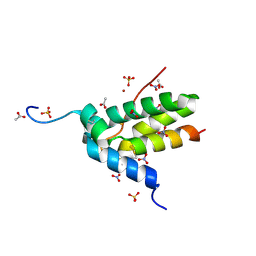 | | Crystal structure of Chitin Binding Protein from Iberis umbellata L. | | Descriptor: | ACETIC ACID, Chitin Binding Protein, LITHIUM ION, ... | | Authors: | Saeed, A, Betzel, C, Brognaro, H, Alves Franca, B, Mehmood, S, Rajaiah Prabhu, P, Ishaq, U, Akrem, A. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of Chitin Binding Protein from Iberis umbellata L.
To be published
|
|
