4R96
 
 | | Structure of a Llama Glama Fab 48A2 against human cMet | | Descriptor: | Llama glama Fab 48A2 against human cMet H chain, Llama glama Fab 48A2 against human cMet L chain | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
1E3F
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-06-14 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E4H
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | GLYCEROL, PENTABROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-04 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E5A
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | 2,4,6-TRIBROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2JTF
 
 | | Solution Structure of the PHF20L1 MBT domain | | Descriptor: | PHD finger protein 20-like 1 | | Authors: | Brockmann, C, Iberg, A.N, Rehbein, K, Diehl, A, Bedford, M.T, Oschkinat, H. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of Histone H4K20 Methyllysine Recognition by the MBT Domain of PHF20L1
To be Published
|
|
3JQY
 
 | | Crystal Structure of the polySia specific acetyltransferase NeuO | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Polysialic acid O-acetyltransferase | | Authors: | Schulz, E.-C, Bergfeld, A, Muehlenhoff, M, Ficner, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO
PLoS ONE, 6, 2011
|
|
1B87
 
 | |
3MEQ
 
 | | Crystal structure of alcohol dehydrogenase from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase, ... | | Authors: | Arakaki, T.L, Staker, B.L, Gardberg, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Brucella melitensis
To be Published
|
|
4XJE
 
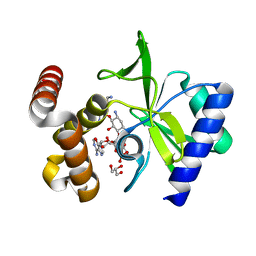 | |
2WHP
 
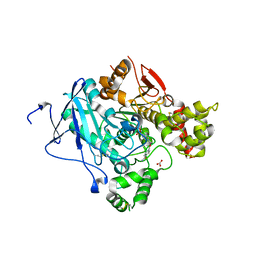 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin and in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
2WHQ
 
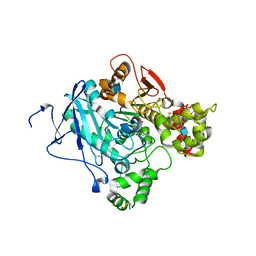 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin (aged) in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
3C92
 
 | | Thermoplasma acidophilum 20S proteasome with a closed gate | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Rabl, J, Smith, D.M, Yu, Y, Chang, S.C, Goldberg, A.L, Cheng, Y. | | Deposit date: | 2008-02-14 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases.
Mol.Cell, 30, 2008
|
|
3FUU
 
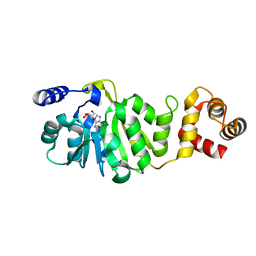 | | T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in complex with Adenosine in space group P212121 | | Descriptor: | ADENOSINE, Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FUX
 
 | | T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in complex with 5'-methylthioadenosine in space group P212121 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FUT
 
 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P21212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3G89
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet and AMP in space group P61 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3FUW
 
 | | T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in complex with 5'-methylthioadenosine in space group P212121 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FUV
 
 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P43212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
1I4N
 
 | | CRYSTAL STRUCTURE OF INDOLEGLYCEROL PHOSPHATE SYNTHASE FROM THERMOTOGA MARITIMA | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Knoechel, T, Pappenberger, A, Jansonius, J.N, Kirschner, K. | | Deposit date: | 2001-02-22 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of indoleglycerol-phosphate synthase from Thermotoga maritima. Kinetic stabilization by salt bridges.
J.Biol.Chem., 277, 2002
|
|
4ZS7
 
 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
4PJV
 
 | | Structure of PARP2 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Edwards, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3G8B
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet in space group I222 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3I0O
 
 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, in complex with ADP and Spectinomcyin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B, Berghuis, A.M. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
2WHR
 
 | | Crystal structure of acetylcholinesterase in complex with K027 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.-G, Schneider, G, Pang, Y.-P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
3F0N
 
 | | Mus Musculus Mevalonate Pyrophosphate Decarboxylase | | Descriptor: | MEVALONATE PYROPHOSPHATE DECARBOXYLASE, PHOSPHATE ION | | Authors: | Walker, J.R, Davis, T, Vesterberg, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Mus Musculus Mevalonate Pyrophosphate Decarboxylase
To be Published
|
|
