8PTQ
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5l | | Descriptor: | 1,2-ETHANEDIOL, Monoglyceride lipase, methyl 4-[(2~{S},3~{R})-3-(4-fluorophenyl)-1-(1-methanoylpiperidin-4-yl)-4-oxidanylidene-azetidin-2-yl]benzoate | | Authors: | Butini, S, Grether, U, Benz, J, Leibrock, L, Maramai, S, Papa, A, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
8PTC
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5d | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3~{R},4~{S})-2-oxidanylidene-3,4-diphenyl-azetidin-1-yl]piperidine-1-carbaldehyde, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
8PTR
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5r | | Descriptor: | (3~{R},4~{S})-4-(1,3-benzodioxol-5-yl)-1-[1-(benzotriazol-1-ylcarbonyl)piperidin-4-yl]-3-(3-fluorophenyl)azetidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
7PRM
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 13 | | Descriptor: | (4~{R})-1-[4-(4-fluorophenyl)phenyl]-4-[4-(furan-2-ylcarbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Grether, U, Gobbi, L, Kuhn, B, Collin, L, Leibrock, L, Heer, D, Wittwer, M, Benz, J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of High Brain-Penetrant and Reversible Monoacylglycerol Lipase PET Tracers for Neuroimaging.
J.Med.Chem., 65, 2022
|
|
7ZPG
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH LIGAND | | Descriptor: | Monoglyceride lipase, [(7R,9aR)-7-(4-chlorophenyl)-1,3,4,6,7,8,9,9a-octahydropyrido[1,2-a]pyrazin-2-yl]-(2-bromanyl-3-methoxy-phenyl)methanone | | Authors: | Kemble, A, Hornsperger, B, Ruf, I, Richter, H, Benz, J, Kuhn, B, Heer, D, Wittwer, M, Engelhardt, B, Grether, U, Collin, L, Leibrock, L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Plos One, 17, 2022
|
|
7BBG
 
 | | CRYSTAL STRUCTURE OF HLA-A2-WT1-RMF AND FAB 11D06 | | Descriptor: | Beta-2-microglobulin, Heavy chain of Fab fragment 11D06, Light chain of Fab fragment 11D06, ... | | Authors: | Bujotzek, A, Georges, G, Hanisch, L.J, Klein, C, Benz, J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Targeting intracellular WT1 in AML with a novel RMF-peptide-MHC-specific T-cell bispecific antibody.
Blood, 138, 2021
|
|
8AQF
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a peripheral restricted, reversible monoacylglycerol lipase inhibitor that reduces liver injury and chemotherapy-induced neuropathy
To Be Published
|
|
1CJA
 
 | | ACTIN-FRAGMIN KINASE, CATALYTIC DOMAIN FROM PHYSARUM POLYCEPHALUM | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN (ACTIN-FRAGMIN KINASE) | | Authors: | Steinbacher, S, Hof, P, Eichinger, L, Schleicher, M, Gettemans, J, Vandekerckhove, J, Huber, R, Benz, J. | | Deposit date: | 1999-04-08 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Physarum polycephalum actin-fragmin kinase: an atypical protein kinase with a specialized substrate-binding domain.
EMBO J., 18, 1999
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XZ9
 
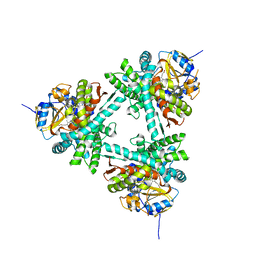 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | Descriptor: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
3HHD
 
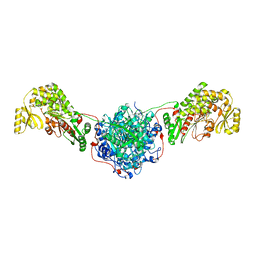 | | Structure of the Human Fatty Acid Synthase KS-MAT Didomain as a Framework for Inhibitor Design. | | Descriptor: | CHLORIDE ION, Fatty acid synthase | | Authors: | Pappenberger, G.M, Benz, J, Thoma, R, Rudolph, M.G. | | Deposit date: | 2009-05-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human fatty acid synthase KS-MAT didomain as a framework for inhibitor design.
J.Mol.Biol., 397, 2010
|
|
6S5A
 
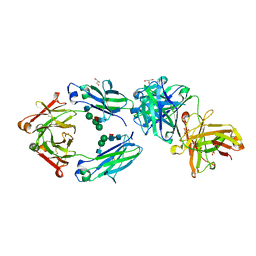 | | CRYSTAL STRUCTURE OF FC P329G LALA WITH ANTI FC P329G FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc P329G LALA, ... | | Authors: | Ehler, A, Darowski, D, Jost, C, Stubenrauch, K, Wessels, U, Benz, J, Birk, M, Freimoser-Grundschober, A, Bruenker, P, Moessner, E, Umana, P, Kobold, S, Klein, C. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | P329G-CAR-J: a novel Jurkat-NFAT-based CAR-T reporter system recognizing the P329G Fc mutation.
Protein Eng.Des.Sel., 32, 2019
|
|
3H52
 
 | | Crystal structure of the antagonist form of human glucocorticoid receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Benz, J, D'Arcy, B, Stihle, M, Burger, D, Thoma, R, Ruf, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations
J.Mol.Biol., 395, 2010
|
|
5IKC
 
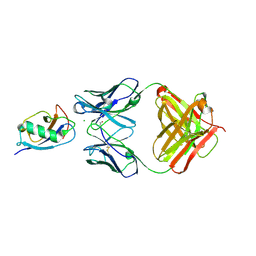 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN in complex with FAB | | Descriptor: | CHLORIDE ION, Ighg protein, MAb 6H10 light chain, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Thoma, R, Rudolph, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IOI
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5IO9
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
4MJO
 
 | | Human liver fructose-1,6-bisphosphatase(d-fructose-1,6-bisphosphate, 1-phosphohydrolase) (e.c.3.1.3.11) complexed with the allosteric inhibitor 3 | | Descriptor: | Fructose-1,6-bisphosphatase 1, N-({4-bromo-6-[(methylcarbamoyl)amino]pyridin-2-yl}carbamoyl)-5-(2-methoxyethyl)-4-methylthiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Tetaz, T, Benz, J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of protein-ligand binding constants of a cooperatively regulated tetrameric enzyme using electrospray mass spectrometry.
Acs Chem.Biol., 9, 2014
|
|
5IN7
 
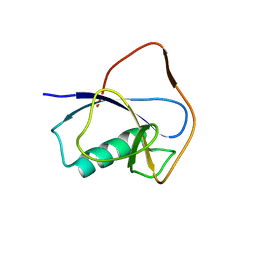 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5N89
 
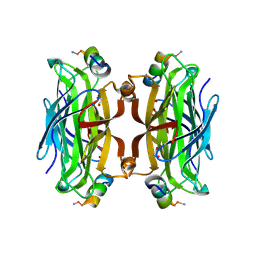 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE GNSFDDWLASKG | | Descriptor: | GLY-ASN-SER-PHE-ASP-ASP-TRP-LEU-ALA-SER-LYS-GLY-NH2, GLYCEROL, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
3S3X
 
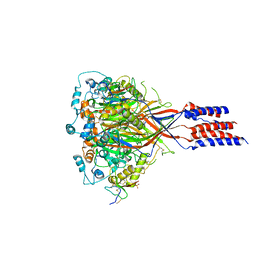 | | Structure of chicken acid-sensing ion channel 1 AT 3.0 A resolution in complex with psalmotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, neuronal, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
5M6Q
 
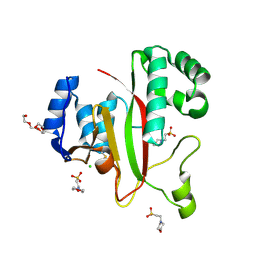 | | Crystal Structure of Kutzneria albida transglutaminase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Steffen, W, Benz, J, Rudolph, M.G. | | Deposit date: | 2016-10-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a microbial transglutaminase enabling highly site-specific labeling of proteins.
J. Biol. Chem., 292, 2017
|
|
4GER
 
 | | Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa | | Descriptor: | CALCIUM ION, Gentlyase metalloprotease, LYSINE, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Schmidt, M, Sobek, H. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5N8W
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN with D-amino acid containing peptide GGwhdeatwkpG | | Descriptor: | GLY-GLY-DTR-DHI-DAS-DGL-DAL-DTH-DTR-DLY, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
