1UXY
 
 | | MURB MUTANT WITH SER 229 REPLACED BY ALA, COMPLEX WITH ENOLPYRUVYL-UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-A resolution.
Biochemistry, 36, 1997
|
|
2P83
 
 | | Potent and selective isophthalamide S2 hydroxyethylamine inhibitor of BACE1 | | Descriptor: | Beta-secretase 1, N~3~-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-N~1~,N~1~-DIPROPYLBENZENE-1,3,5-TRICARBOXAMIDE, PHOSPHATE ION | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2007-03-21 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent and selective isophthalamide S(2) hydroxyethylamine inhibitors of BACE1.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1MBB
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-3(N-ACETYLGLUCOSAMINYL)BUTYRIC ACID | | Authors: | Benson, T.E, Lees, W.J, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | (E)-enolbutyryl-UDP-N-acetylglucosamine as a mechanistic probe of UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 35, 1996
|
|
1MBT
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls.
Structure, 4, 1996
|
|
2MBR
 
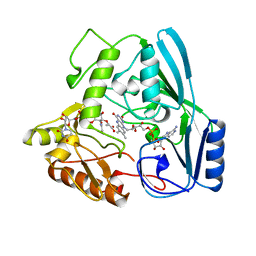 | | MURB WILD TYPE, COMPLEX WITH ENOLPYRUVYL-UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-A resolution.
Biochemistry, 36, 1997
|
|
1LRZ
 
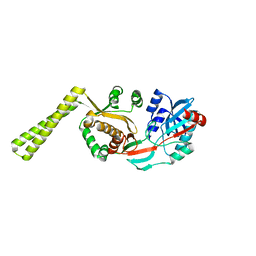 | | x-ray crystal structure of staphylococcus aureus femA | | Descriptor: | factor essential for expression of methicillin resistance | | Authors: | Benson, T, Prince, D, Mutchler, V, Curry, K, Ho, A, Sarver, R, Hagadorn, J, Choi, G, Garlick, R. | | Deposit date: | 2002-05-16 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of Staphylococcus aureus FemA.
Structure, 10, 2002
|
|
2IQG
 
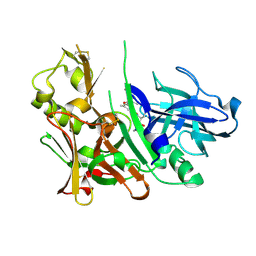 | | Crystal Structure of Hydroxyethyl Secondary Amine-based Peptidomimetic Inhibitor of Human Beta-Secretase (BACE) | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-IODOBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Benson, T.E, Woods, D.D, Prince, D.B, Tomasselli, A.G, Emmons, T.L. | | Deposit date: | 2006-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Crystal Structure of Hydroxyethyl Secondary Amine-Based Peptidomimetic Inhibitors of Human beta-Secretase.
J.Med.Chem., 50, 2007
|
|
2HIZ
 
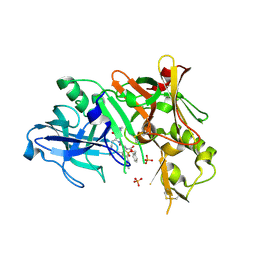 | | Crystal Structure of human beta-secretase (BACE) in the presence of an inhibitor | | Descriptor: | BENZYL [(1S)-2-({(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}AMINO)-2-OXO-1-{[(1-PROPYLBUTYL)SULFONYL]METHYL}ETHYL]CARBAMATE, Beta-secretase 1, PHOSPHATE ION | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of potent inhibitors of human beta-secretase. Part 1.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2HM1
 
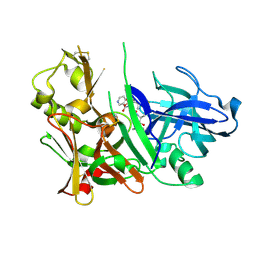 | | Crystal Structure of human beta-secretase (BACE) in the presence of an inhibitor (2) | | Descriptor: | Beta-secretase 1, N-{(1S)-2-({(1S,2R)-1-(3,5-DIFLUOROBENZYL)-3-[(3-ETHYLBENZYL)AMINO]-2-HYDROXYPROPYL}AMINO)-2-OXO-1-[(PENTYLSULFONYL)METHYL]ETHYL}NICOTINAMIDE | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2006-07-10 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent inhibitors of human beta-secretase. Part 2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1HSK
 
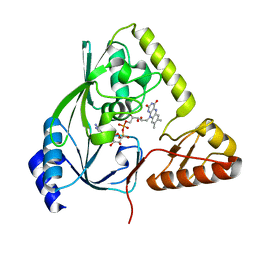 | | CRYSTAL STRUCTURE OF S. AUREUS MURB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-ACETYLENOLPYRUVOYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Harris, M.S, Choi, G.H, Cialdella, J.I, Herberg, J.T, Martin Jr, J.P, Baldwin, E.T. | | Deposit date: | 2000-12-27 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural variation for MurB: X-ray crystal structure of Staphylococcus aureus UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 40, 2001
|
|
5K5M
 
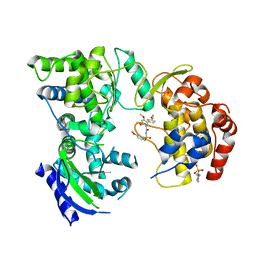 | |
3LXN
 
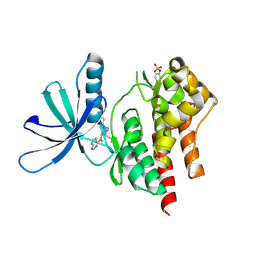 | |
3LXP
 
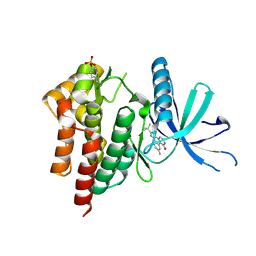 | |
6XD1
 
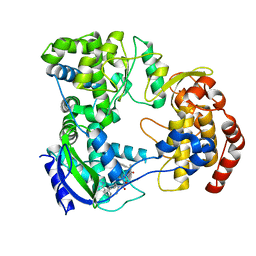 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-640 | | Descriptor: | (2R)-4-(butyl{[2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}carbamoyl)-1-(2,2-diphenylpropanoyl)piperazine-2-carboxylic acid, RNA-dependent RNA polymerase, ZINC ION | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
6XD0
 
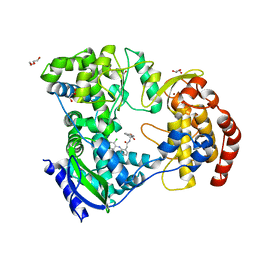 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-434 | | Descriptor: | 2-[({2-[(2,6-dichlorophenyl)amino]phenyl}acetyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
6OB0
 
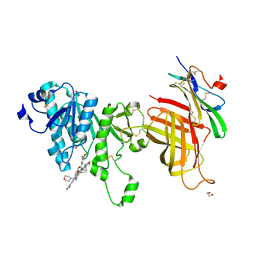 | | Compound 2 bound structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OAU
 
 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in GnTI-deficient HEK293-F cells | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OAZ
 
 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3LXK
 
 | | Structural and Thermodynamic Characterization of the TYK2 and JAK3 Kinase Domains in Complex with CP-690550 and CMP-6 | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Tyrosine-protein kinase JAK3 | | Authors: | Chrencik, J.E, Patny, A, Leung, I.K, Korniski, B, Emmons, T.L, Benson, T.E. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6.
J.Mol.Biol., 400, 2010
|
|
3LXL
 
 | |
1A1J
 
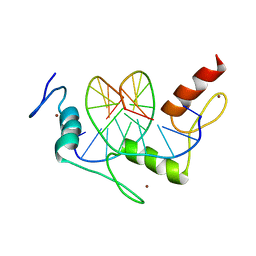 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (RADR ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1G
 
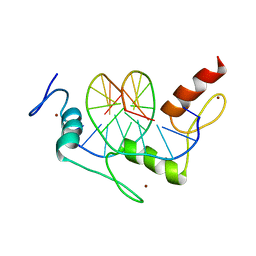 | | DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), DSNR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1F
 
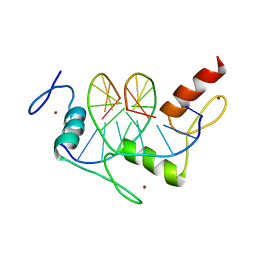 | | DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), THREE-FINGER ZIF268 PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1H
 
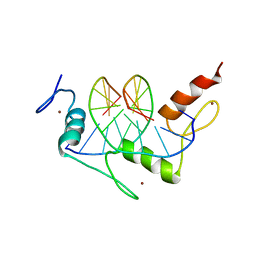 | | QGSR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), QGSR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1I
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
