6TSF
 
 | | Crystal structure of human L ferritin (HuLf) Fe(III)-loaded for 60 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
5WJI
 
 | | Crystal structure of the F61S mutant of HsNUDT16 | | Descriptor: | ACETIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-07-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
6TS0
 
 | | Crystal structure of human L ferritin (HuLf) triple variant E60A-E61A-E64A Fe(III)-loaded for 30 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
4YY6
 
 | | Crystal structure of BRD9 Bromodomain bound to a butyryllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4YYD
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6BUU
 
 | | Crystal structure of AKT1 (aa 144-480) with a bisubstrate | | Descriptor: | GLY-ARG-PRO-ARG-THR-THR-ZXW-PHE-ALA-GLU, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Gabelli, S.B, Cole, P.A. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-22 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
6TS1
 
 | | Crystal structure of human L ferritin (HuLf) triple variant E60A-E61A-E64A Fe(III)-loaded for 60 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TR9
 
 | | Crystal structure of human L ferritin (HuLf) triple variant E60A-E61A-E64A | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TSJ
 
 | | Crystal structure of human L ferritin (HuLf) Fe(III)-loaded for 15 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6ALA
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5WJA
 
 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WKW
 
 | | Crystal structure of apo wild type peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WM0
 
 | |
6UUF
 
 | | Crystal structure of a Nudix Hydrolase from M. Smegmatis, RenU | | Descriptor: | Nudix Hydrolase, RenU | | Authors: | Wright, K.M, Yoder, J, Shoemaker, S, Hernandez, A, Iheanacho, A, Marques, I, Amzel, M.L, Gabelli, S.B. | | Deposit date: | 2019-10-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of RenU
To Be Published
|
|
6VO7
 
 | |
7UTT
 
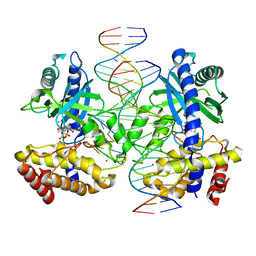 | |
7UXW
 
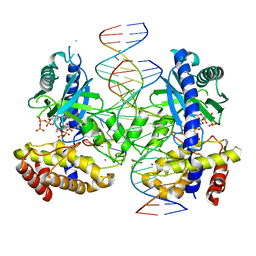 | | Structure of ATP and GTP bind to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYQ
 
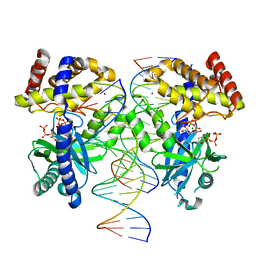 | | Structure of GTP binds to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYZ
 
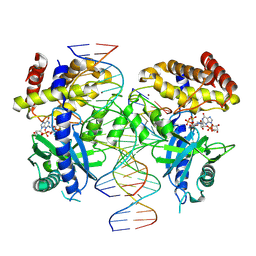 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UZR
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, MANGANESE (II) ION, Palindromic DNA18, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
3CCN
 
 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | Descriptor: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | Authors: | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
5W6X
 
 | |
5VY2
 
 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5V84
 
 | | CECR2 in complex with Cpd6 (6-allyl-N,2-dimethyl-7-oxo-N-(1-(1-phenylethyl)piperidin-4-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4-carboxamide) | | Descriptor: | Cat eye syndrome critical region protein 2, N,2-dimethyl-7-oxo-N-{1-[(1S)-1-phenylethyl]piperidin-4-yl}-6-(prop-2-en-1-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4- carboxamide, SULFATE ION | | Authors: | Murray, J.M, Kiefer, J.R, Jayaran, H, Bellon, S, Boy, F. | | Deposit date: | 2017-03-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | GNE-886: A Potent and Selective Inhibitor of the Cat Eye Syndrome Chromosome Region Candidate 2 Bromodomain (CECR2).
ACS Med Chem Lett, 8, 2017
|
|
