5VI5
 
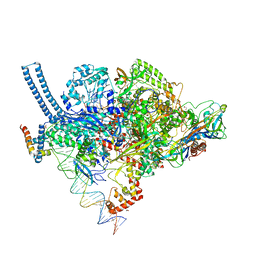 | | Structure of Mycobacterium smegmatis transcription initiation complex with a full transcription bubble | | Descriptor: | 1,2-ETHANEDIOL, DNA (44-MER), DNA (49-MER), ... | | Authors: | Darst, S.A, Campbell, E.A, Lilic, M. | | Deposit date: | 2017-04-14 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures.
Nat Commun, 8, 2017
|
|
6OUL
 
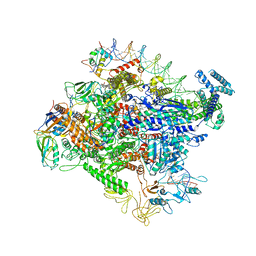 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound to rpsTP2 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
7JKA
 
 | |
7JIZ
 
 | |
3MLQ
 
 | |
6P1K
 
 | | Cryo-EM structure of Escherichia coli sigma70 bound RNAP polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
4XAY
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R8, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD3
 
 | | Phosphotriesterase variant E3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD6
 
 | | Phosphotriesterase Variant E2a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E2, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAF
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
6PSU
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi2) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSR
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSW
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPo) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSS
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5a) with TraR and mutant rpsT P2 promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSQ
 
 | | Escherichia coli RNA polymerase closed complex (TRPc) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
8SQ9
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQJ
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQK
 
 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8DY7
 
 | |
8DY9
 
 | |
6PST
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5b) with TraR and mutant rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
2Z2S
 
 | |
3TBI
 
 | |
