8OXJ
 
 | |
5MQV
 
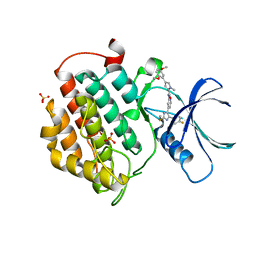 | | Crystal structure of human Casein Kinase I delta in complex with 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide | | Descriptor: | 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
2QUB
 
 | |
2QUA
 
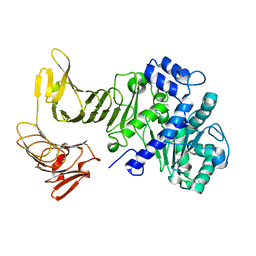 | | Crystal structure of LipA from Serratia marcescens | | Descriptor: | CALCIUM ION, Extracellular lipase | | Authors: | Meier, R, Baumann, U. | | Deposit date: | 2007-08-04 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A calcium-gated lid and a large beta-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens.
J.Biol.Chem., 282, 2007
|
|
3HB2
 
 | | PrtC methionine mutants: M226I | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-04 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3HDA
 
 | | PrtC methionine mutants: M226A_DESY | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3HBU
 
 | | PrtC methionine mutants: M226H DESY | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3HBV
 
 | | PrtC methionine mutants: M226A in-house | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-05 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
8ADB
 
 | | Viral tegument-like DUBs | | Descriptor: | CITRIC ACID, Polyubiquitin-C, Wc-VDT1, ... | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
8ADC
 
 | | Viral tegument-like DUBs | | Descriptor: | Viral deubiquitinating enzyme | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
2IU6
 
 | | REGULATION OF THE DHA OPERON OF LACTOCOCCUS LACTIS | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCEROL | | Authors: | Srinivas, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2006-05-27 | | Release date: | 2006-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regulation of the Dha Operon of Lactococcus Lactis: A Deviation from the Rule Followed by the Tetr Family of Transcription Regulators
J.Biol.Chem., 281, 2006
|
|
2IU4
 
 | | Dihydroxyacetone kinase operon co-activator Dha-DhaQ | | Descriptor: | DIHYDROXYACETONE KINASE, SULFATE ION | | Authors: | Srinivas, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2006-05-27 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Regulation of the Dha Operon of Lactococcus Lactis: A Deviation from the Rule Followed by the Tetr Family of Transcription Regulators
J.Biol.Chem., 281, 2006
|
|
8CMR
 
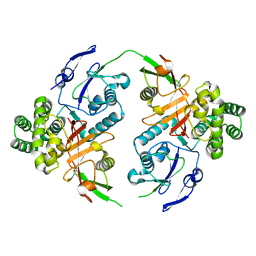 | | Linear specific OTU-type DUB SnOTU from the pathogen S. negenvensis in complex with linear di-ubiquitin | | Descriptor: | OTU domain-containing protein, Polyubiquitin-B | | Authors: | Uthoff, M, Hermanns, T, Boll, V, Hofmann, K, Baumann, U. | | Deposit date: | 2023-02-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural diversity in deubiquitinases of the Chlamydia-like bacterium Simkania negevensis.
Nat Commun, 14, 2023
|
|
5FR2
 
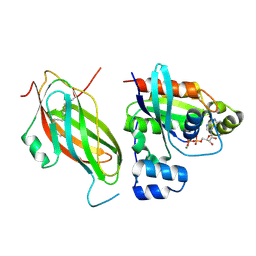 | | Farnesylated RhoA-GDP in complex with RhoGDI-alpha, lysine acetylated at K178 | | Descriptor: | FARNESYL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuhlmann, N, Wroblowski, S, Knyphausen, P, de Boor, S, Brenig, J, Zienert, A.Y, Meyer-Teschendorf, K, Praefcke, G.J.K, Nolte, H, Krueger, M, Schacherl, M, Baumann, U, James, L.C, Chin, J.W, Lammers, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural and Mechanistic Insights Into the Regulation of the Fundamental Rho-Regulator Rhogdi Alpha by Lysine Acetylation.
J.Biol.Chem., 291, 2016
|
|
5FYQ
 
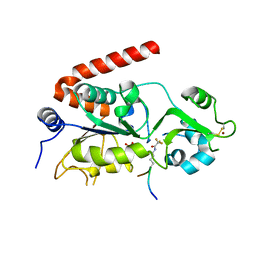 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
5G5R
 
 | |
2BTD
 
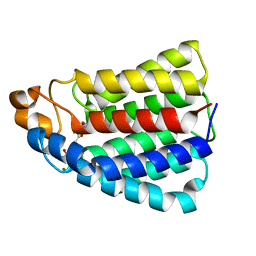 | | Crystal structure of DhaL from E. coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-DEPENDENT DIHYDROXYACETONE KINASE | | Authors: | Oberholzer, A.E, Schneider, P, Bachler, C, Baumann, U, Erni, B. | | Deposit date: | 2005-05-27 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Nucleotide-Binding Subunit Dhal of the Escherichia Coli Dihydroxyacetone Kinase.
J.Mol.Biol., 359, 2006
|
|
2BG5
 
 | | Crystal Structure of the Phosphoenolpyruvate-binding Enzyme I-Domain from the Thermoanaerobacter tengcongensis PEP: Sugar Phosphotransferase System (PTS) | | Descriptor: | PHOSPHOENOLPYRUVATE-PROTEIN KINASE | | Authors: | Oberholzer, A.E, Bumann, M, Schneider, P, Baechler, C, Siebold, C, Baumann, U, Erni, B. | | Deposit date: | 2004-12-17 | | Release date: | 2005-02-02 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of the Phosphoenolpyruvate-Binding Enzyme I-Domain from the Thermoanaerobacter Tengcongensis Pep: Sugar Phosphotransferase System (Pts)
J.Mol.Biol., 346, 2005
|
|
5G5X
 
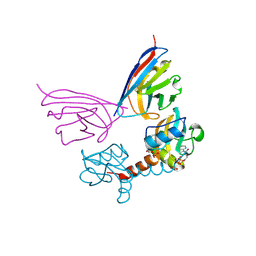 | |
2CE7
 
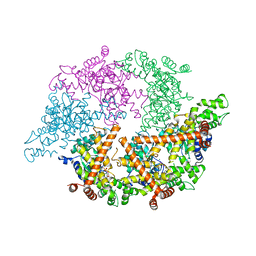 | | EDTA treated | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION PROTEIN FTSH, MAGNESIUM ION, ... | | Authors: | Bieniossek, C, Baumann, U. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Molecular Architecture of the Metalloprotease Ftsh.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CKI
 
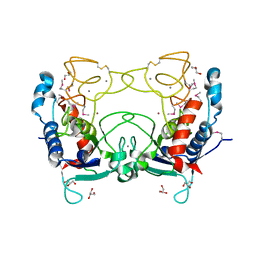 | | Structure of Ulilysin, a member of the pappalysin family of metzincin metalloendopeptidases. | | Descriptor: | ARGININE, CALCIUM ION, GLYCEROL, ... | | Authors: | Tallant, C, Garcia-Castellanos, R, Seco, J, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2006-04-19 | | Release date: | 2006-05-09 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Analysis of Ulilysin, the Structural Prototype of a New Family of Metzincin Metalloproteases.
J.Biol.Chem., 281, 2006
|
|
7OIY
 
 | | Crystal structure of the ZUFSP family member Mug105 | | Descriptor: | SODIUM ION, Ubiquitin carboxyl-terminal hydrolase mug105 | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
7OJE
 
 | | Crystal structure of the covalent complex between Tribolium castaneum deubiquitinase ZUP and Ubiquitin-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Lys-63-specific deubiquitinase ZUFSP, ... | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
1JIW
 
 | | Crystal structure of the APR-APRin complex | | Descriptor: | ALKALINE METALLOPROTEINASE, CALCIUM ION, PROTEINASE INHIBITOR, ... | | Authors: | Hege, T, Feltzer, R.E, Gray, R.D, Baumann, U. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a complex between Pseudomonas aeruginosa alkaline protease and its cognate inhibitor: inhibition by a zinc-NH2 coordinative bond
J.Biol.Chem., 276, 2001
|
|
5A0P
 
 | | Apo-structure of metalloprotease Zmp1 from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
