1SQL
 
 | | Crystal structure of 7,8-dihydroneopterin aldolase in complex with guanine | | Descriptor: | GUANINE, dihydroneopterin aldolase | | Authors: | Bauer, S, Schott, A.K, Illarionova, V, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2004-03-19 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Tetrahydrofolate in Plants: Crystal Structure of 7,8-Dihydroneopterin Aldolase from Arabidopsis thaliana Reveals a Novel Adolase Class.
J.Mol.Biol., 339, 2004
|
|
1N05
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | putative Riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1N06
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PUTATIVE riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1N08
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ZINC ION, putative riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
1N07
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, PUTATIVE riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
4YM9
 
 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the novel inhibitor JM102 | | Descriptor: | 2-ethyl-2-(hydroxymethyl)-N-(6-methylpyridin-3-yl)butanamide, ACETATE ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Hofbauer, S, Brito, J.A, Mulchande, J, Nogly, P, Pessanha, M, Moreira, R, Archer, M. | | Deposit date: | 2015-03-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clickable 4-Oxo-beta-lactam-Based Selective Probing for Human Neutrophil Elastase Related Proteomes.
ChemMedChem, 11, 2016
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6SV3
 
 | | Structure of coproheme-LmCpfC | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Ferrochelatase, GLYCEROL | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64000869 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
6FXJ
 
 | | Structure of coproheme decarboxylase from Listeria monocytogenes in complex with iron coproporphyrin III | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, CHLORIDE ION, N-PROPANOL, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
6FXQ
 
 | | Structure of coproheme decarboxylase from Listeria monocytogenes during turnover | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G, Puehringer, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
1HQ8
 
 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
6HIJ
 
 | | Cryo-EM structure of the human ABCG2-MZ29-Fab complex with cholesterol and PE lipids docked | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ATP-binding cassette sub-family G member 2, CHOLESTEROL, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-08-30 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat.Struct.Mol.Biol., 25, 2018
|
|
1B3J
 
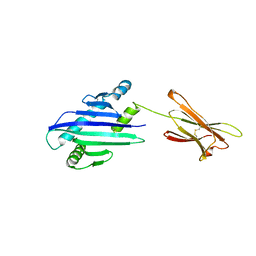 | | STRUCTURE OF THE MHC CLASS I HOMOLOG MIC-A, A GAMMADELTA T CELL LIGAND | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I HOMOLOG MIC-A | | Authors: | Li, P, Willie, S, Bauer, S, Morris, D, Spies, T, Strong, R. | | Deposit date: | 1998-12-11 | | Release date: | 1999-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand.
Immunity, 10, 1999
|
|
3GJE
 
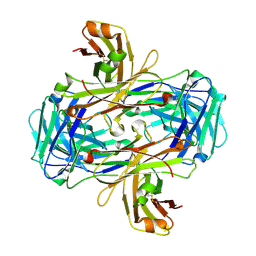 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Fab Heavy Chain, Fab Light Chain | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GJF
 
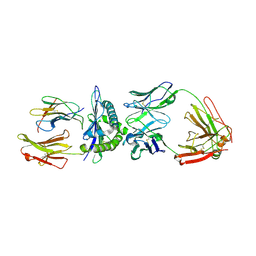 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Antibody heavy chain, Antibody light chain, Beta-2-microglobulin, ... | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2Y8I
 
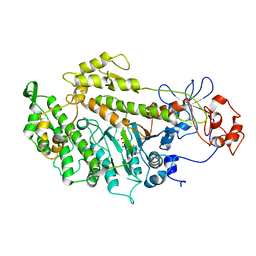 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
2Y9E
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.397 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
2Y0R
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
6FFC
 
 | | Structure of an inhibitor-bound ABC transporter | | Descriptor: | ATP-binding cassette sub-family G member 2, ~{tert}-butyl 3-[(2~{S},5~{S},8~{S})-14-cyclopentyloxy-2-(2-methylpropyl)-4,7-bis(oxidanylidene)-3,6,17-triazatetracyclo[8.7.0.0^{3,8}.0^{11,16}]heptadeca-1(10),11,13,15-tetraen-5-yl]propanoate | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3MJX
 
 | | Crystal structure of myosin-2 motor domain in complex with ADP-Metavanadate and blebbistatin | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Fedorov, R, Baruch, P, Bauer, S, Manstein, D.J. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of pentabromopseudilin inhibition of myosin motor activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1TKU
 
 | | Crystal Structure of 3,4-Dihydroxy-2-butanone 4-phosphate Synthase of Candida albicans in complex with Ribulose-5-phosphate | | Descriptor: | 3,4-Dihydroxy-2-butanone 4-phosphate Synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Echt, S, Bauer, S, Steinbacher, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-06-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans.
J.Mol.Biol., 341, 2004
|
|
1TKS
 
 | | Crystal structure of 3,4-Dihydroxy-2-butanone 4-phosphate Synthase of Candida albicans | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase | | Authors: | Echt, S, Bauer, S, Steinbacher, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-06-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans.
J.Mol.Biol., 341, 2004
|
|
8BBV
 
 | | Coproporphyrin III - LmCpfC complex soaked 2min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Coproporphyrin III ferrochelatase, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
8AW7
 
 | | Structure of coproporphyrin III-LmCpfC R45L | | Descriptor: | Coproporphyrin III ferrochelatase, GLYCEROL, coproporphyrin III | | Authors: | Gabler, T, Hofbauer, S, Pfanzagl, V. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
8AT8
 
 | | Structure of coproporphyrin III-LmCpfC | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
