2QE2
 
 | | Structure of HCV NS5B Bound to an Anthranilic Acid Inhibitor | | Descriptor: | 2-{[N-(2-ACETYL-5-CHLORO-4-FLUOROPHENYL)GLYCYL]AMINO}BENZOIC ACID, RNA-directed RNA polymerase | | Authors: | Chopra, R, Svenson, K, Bard, J. | | Deposit date: | 2007-06-22 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Anthranilic Acid Derivatives as a Novel Class of Allosteric Inhibitors of Hepatitis C NS5B Polymerase
J.Med.Chem., 50, 2007
|
|
5WO3
 
 | | Chaperone Spy bound to Im7 (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WO2
 
 | | Chaperone Spy bound to Casein Fragment (Casein un-modeled) | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Periplasmic chaperone Spy, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5KCI
 
 | | Crystal Structure of HTC1 | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein YPL067C, ... | | Authors: | Martin, R.M, Horowitz, S, Koepnick, B, Cooper, S, Flatten, J, Rogawski, D.S, Koropatkin, N.M, Beinlich, F.R.M, Players, F, Students, U.M, Popovic, Z, Baker, D, Khatib, F, Bardwell, J.C.A. | | Deposit date: | 2016-06-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Determining crystal structures through crowdsourcing and coursework.
Nat Commun, 7, 2016
|
|
8BV3
 
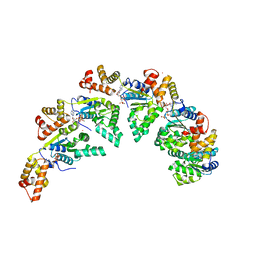 | | Bacillus subtilis DnaA domain III structure | | Descriptor: | Chromosomal replication initiator protein DnaA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pintar, S, Hubbard, J.A. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The bacterial replication origin BUS promotes nucleobase capture.
Nat Commun, 14, 2023
|
|
7TLX
 
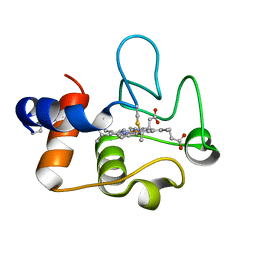 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
4TT4
 
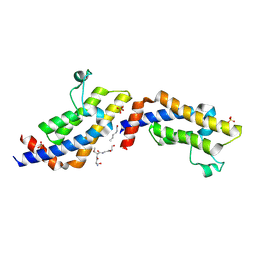 | | Crystal structure of ATAD2A bromodomain complexed with H3(1-21)K14Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, Histone H3(1-21)K4Ac, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT2
 
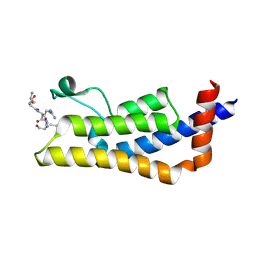 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU6
 
 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU4
 
 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT6
 
 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
8DSV
 
 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
 | |
8DQ8
 
 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
7TQR
 
 | |
5WO1
 
 | | Chaperone Spy H96L bound to Im7 L18A L19A L37A (Im7 un-modeled) | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, IMIDAZOLE, ... | | Authors: | Horowitz, S, Koldewey, P, Martin, R, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5ISO
 
 | | STRUCTURE OF FULL LENGTH HUMAN AMPK (NON-PHOSPHORYLATED AT T-LOOP) IN COMPLEX WITH A SMALL MOLECULE ACTIVATOR, A BENZIMIDAZOLE DERIVATIVE (991) | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Xiao, B, Hubbard, J.A, Gamblin, S.J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | STRUCTURE OF FULL LENGTH HUMAN AMPK (NON-PHOSPHORYLATED AT T-LOOP) IN COMPLEX WITH A SMALL MOLECULE ACTIVATOR, A BENZIMIDAZOLE DERIVATIVE (991)
To Be Published
|
|
6X84
 
 | |
1ZCP
 
 | |
2O20
 
 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | Descriptor: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | Authors: | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6OWZ
 
 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWX
 
 | | Spy H96L:Im7 L18pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWY
 
 | | Spy H96L:Im7 K20pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
