1F5Z
 
 | |
1F6P
 
 | |
1KAR
 
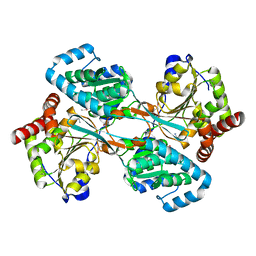 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH HISTAMINE (INHIBITOR), ZINC AND NAD (COFACTOR) | | Descriptor: | HISTAMINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAE
 
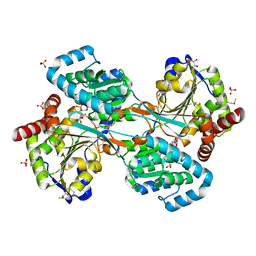 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINOL (SUBSTRATE), ZINC AND NAD (COFACTOR) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Histidinol dehydrogenase, ... | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-01 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K75
 
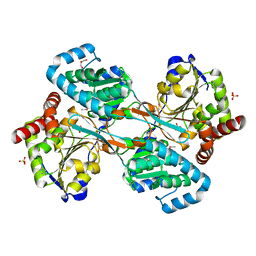 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAH
 
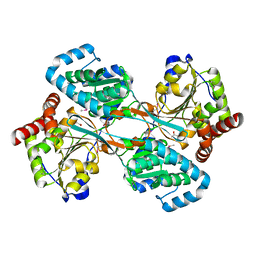 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINE (PRODUCT), ZN AND NAD (COFACTOR) | | Descriptor: | HISTIDINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1F74
 
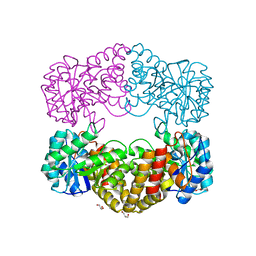 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II COMPLEXED WITH 4-DEOXY-SIALIC ACID | | Descriptor: | 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-2-OXONONANOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F7B
 
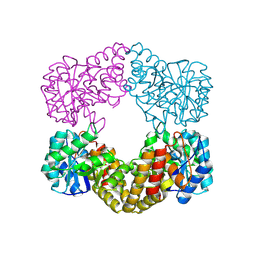 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II IN COMPLEX WITH 4-OXO-SIALIC ACID | | Descriptor: | 4,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDONONANOIC ACID, 6,7,8,9-TETRAHYDROXY-5-METHYLCARBOXAMIDO-4-OXONONANOIC ACID, CHLORIDE ION, ... | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-26 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F73
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM III IN COMPLEX WITH SIALIC ACID ALDITOL | | Descriptor: | 2,4,6,7,8,9-HEXAHYDROXY-5-METHYLCARBOXAMIDO NONANOIC ACID, GLYCEROL, N-ACETYL NEURAMINATE LYASE | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-25 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F6K
 
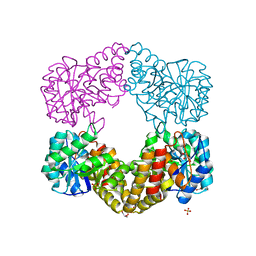 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II | | Descriptor: | GLYCEROL, N-ACETYLNEURAMINATE LYASE, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
5JY5
 
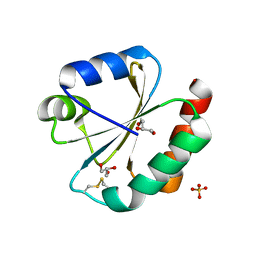 | | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Bravo-Chaucanes, C.P, Abadio, A.K.R, Kioshima, E.S, Felipe, M.S.S, Barbosa, J.A.R.G. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thioredoxin 1 from Cryptococcus neoformans at 1.8 Angstroms resolution
To Be Published
|
|
4JJM
 
 | | Structure of a cyclophilin from Citrus sinensis (CsCyp) in complex with cyclosporin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, cyclosporin A | | Authors: | Campos, B.M, Ambrosio, A.L.B, Souza, T.A.C.B, Barbosa, J.A.R.G, Benedetti, C.E. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A redox 2-cys mechanism regulates the catalytic activity of divergent cyclophilins.
Plant Physiol., 162, 2013
|
|
2B7H
 
 | | Hemoglobin from Cerdocyon thous, a canidae from Brazil, at 2.2 Angstroms resolution | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, hemoglobin alpha chain, ... | | Authors: | Esteves, G.F, Silva, V.C, Bloch Jr, C, Medrano, F.J, Barbosa, J.A.R.G, Freitas, S.M. | | Deposit date: | 2005-10-04 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and biophysical characterization of the Cerdocyon thous, a Canidae from Brazil.
To be Published
|
|
3CB7
 
 | | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang. | | Descriptor: | ACETIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Cancado, F.C, Valerio, A.A, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2008-02-21 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystallographic structure of the digestive lysozyme 2 from Musca domestica at 1.9 Ang.
To be Published
|
|
3KSX
 
 | |
3H79
 
 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
3KSJ
 
 | |
3GZG
 
 | |
6AWE
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2-fluoro-6-(2-pyridin-2-ylethylamino)benzonitrile | | Descriptor: | 2-fluoro-6-{[2-(pyridin-2-yl)ethyl]amino}benzonitrile, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2-fluoro-6-(2-pyridin-2-ylethylamino)benzonitrile
To Be Published
|
|
6B71
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with3-(4-chlorophenyl)-5H,6H-imidazo[2,1-b][1,3]thiazole | | Descriptor: | 3-(4-chlorophenyl)-5,6-dihydroimidazo[2,1-b][1,3]thiazole, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 3-(4-chlorophenyl)-5H,6H-imidazo[2,1-b][1,3]thiazole
To Be Published
|
|
3E4R
 
 | | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nitrate transport protein | | Authors: | Balan, A, Araujo, F.T, Sanches, M, Chirgadze, D.Y, Blundell, T.B, Barbosa, J.A.R.G. | | Deposit date: | 2008-08-12 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES
To be Published
|
|
6MRQ
 
 | | Structure of ToPI1 inhibitor from Tityus obscurus scorpion venom in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Fernandes, J.C, Mourao, C.B.F, Schwartz, E.F, Barbosa, J.A.R.G. | | Deposit date: | 2018-10-15 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Head-to-Tail Cyclization after Interaction with Trypsin: A Scorpion Venom Peptide that Resembles Plant Cyclotides.
J.Med.Chem., 63, 2020
|
|
1NNS
 
 | | L-asparaginase of E. coli in C2 space group and 1.95 A resolution | | Descriptor: | ASPARTIC ACID, L-asparaginase II | | Authors: | Sanches, M, Barbosa, J.A.R.G, de Oliveira, R.T, Neto, J.A.A, Polikarpov, I. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of Escherichia coli L-asparaginase in two monoclinic space groups.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N3B
 
 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
3RU4
 
 | | Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, Bowman-Birk type seed trypsin and chymotrypsin inhibitor, ... | | Authors: | Esteves, G.F, Santos, C.R, Ventura, M.M, Barbosa, J.A.R.G, Freitas, S.M. | | Deposit date: | 2011-05-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of the Bowman-Birk serine protease inhibitor BTCI in complex with trypsin and chymotrypsin
To be Published
|
|
