5X41
 
 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X3X
 
 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
4RFS
 
 | | Structure of a pantothenate energy coupling factor transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Zhang, M, Bao, Z, Zhao, Q, Guo, H, Xu, K, Zhang, P. | | Deposit date: | 2014-09-27 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4II5
 
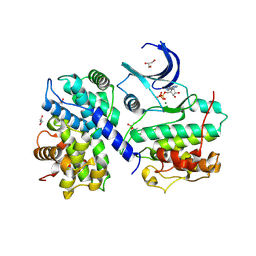 | | Structure of PCDK2/CYCLINA bound to ADP and 1 MAGNESIUM ION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Jacobsen, D.M, Bao, Z.-Q, O'Brien, P.J, Brooks III, C.L, Young, M.A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Price to be paid for two-metal catalysis: Magnesium ions that accelerate chemistry unavoidably limit product release from a PROTEIN KINASE
J.Am.Chem.Soc., 134, 2012
|
|
4I3Z
 
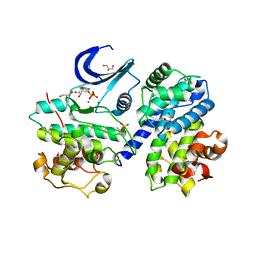 | | Structure of pCDK2/CyclinA bound to ADP and 2 Magnesium ions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Cyclin-A2, ... | | Authors: | Jacobsen, D.M, Bao, Z.-Q, O'Brien, P.J, Brooks, C.L, Young, M.A. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Price To Be Paid for Two-Metal Catalysis: Magnesium Ions That Accelerate Chemistry Unavoidably Limit Product Release from a Protein Kinase
J.Am.Chem.Soc., 134, 2012
|
|
4PVC
 
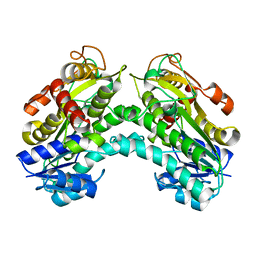 | |
4PVD
 
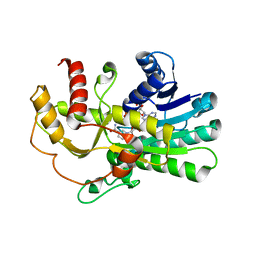 | | Crystal structure of yeast methylglyoxal/isovaleraldehyde reductase Gre2 complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent methylglyoxal reductase GRE2 | | Authors: | Guo, P.C, Bao, Z.Z, Li, W.F, Zhou, C.Z. | | Deposit date: | 2014-03-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast methylglyoxal/isovaleraldehyde reductase Gre2
Biochim.Biophys.Acta, 1844, 2014
|
|
3QHR
 
 | | Structure of a pCDK2/CyclinA transition-state mimic | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CDK2 substrate peptide: PKTPKKAKKL, CHLORIDE ION, ... | | Authors: | Young, M.A, Jacobsen, D.M, Bao, Z.Q. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Briefly Bound to Activate: Transient Binding of a Second Catalytic Magnesium Activates the Structure and Dynamics of CDK2 Kinase for Catalysis.
Structure, 19, 2011
|
|
3QHW
 
 | | Structure of a pCDK2/CyclinA transition-state mimic | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CDK2 substrate peptide: PKTPKKAKKL, ... | | Authors: | Young, M.A, Jacobsen, D.M, Bao, Z.Q. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Briefly Bound to Activate: Transient Binding of a Second Catalytic Magnesium Activates the Structure and Dynamics of CDK2 Kinase for Catalysis.
Structure, 19, 2011
|
|
3QWB
 
 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 complexed with NADPH | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QWA
 
 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 | | Descriptor: | Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
7FAC
 
 | | Crystal Structure of C-terminus of the non-structural protein 2 from SARS coronavirus | | Descriptor: | Non-structural protein 2, ZINC ION | | Authors: | Li, Y.Y, Ren, Z.L, Bao, Z.H, Ming, Z.H, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2021-07-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function.
Annu.Rev.Biochem., 91, 2022
|
|
7FA1
 
 | | Crystal Structure of N-terminus of the non-structural protein 2 from SARS coronavirus | | Descriptor: | Non-structural protein 2, ZINC ION | | Authors: | Li, Y.Y, Ren, Z.L, Bao, Z.H, Ming, Z.H, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2021-07-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function.
Annu.Rev.Biochem., 91, 2022
|
|
6X5A
 
 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6X59
 
 | | The mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA, Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
2FTD
 
 | | Crystal structure of Cathepsin K complexed with 7-Methyl-Substituted Azepan-3-one compound | | Descriptor: | Cathepsin K, N-[(1S)-1-({[(3S,4S,7R)-3-HYDROXY-7-METHYL-1-(PYRIDIN-2-YLSULFONYL)-2,3,4,7-TETRAHYDRO-1H-AZEPIN-4-YL]AMINO}CARBONYL)-3-METHYLBUTYL]-1-BENZOFURAN-2-CARBOXAMIDE | | Authors: | Yamashita, D.S, Baoguang, Z. | | Deposit date: | 2006-01-24 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure activity relationships of 5-, 6-, and 7-methyl-substituted azepan-3-one cathepsin K inhibitors.
J.Med.Chem., 49, 2006
|
|
