3CG6
 
 | | Crystal structure of Gadd45 gamma | | Descriptor: | Growth arrest and DNA-damage-inducible 45 gamma | | Authors: | Schrag, J.D, Jiralerspong, S, Banville, M, Jaramillo, M.L, O'Connor-McCourt, M.D. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure and dimerization interface of GADD45gamma.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5A6W
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
3MEZ
 
 | | X-ray structural analysis of a mannose specific lectin from dutch crocus (crocus vernus) | | Descriptor: | FORMIC ACID, GLYCEROL, Mannose-specific lectin 3 chain 1, ... | | Authors: | Akrem, A, Meyer, A, Perbandt, M, Voelter, W, Buck, F, Betzel, C. | | Deposit date: | 2010-04-01 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray structural analysis of a mannose specific lectin from dutch crocus (crocus vernus)
To be Published
|
|
3GVN
 
 | | The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites | | Descriptor: | 5'-R(*CP*CP*UP*CP*AP*CP*C)-3', 5'-R(*GP*GP*UP*GP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Eichert, A, Furste, J.P, Schreiber, A, Perbandt, M, Betzel, C, Erdmann, V.A, Forster, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
5OD4
 
 | |
2AYW
 
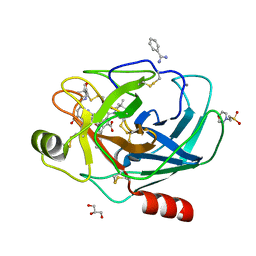 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3ZVG
 
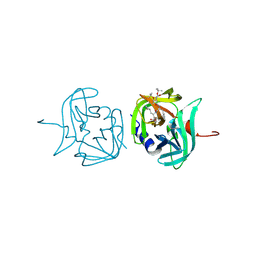 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | Descriptor: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVD
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 83 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
1E3S
 
 | | Rat brain 3-hydroxyacyl-CoA dehydrogenase binary complex with NADH | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Powell, A.J, Read, J.A, Banfield, M.J, Brady, R.L. | | Deposit date: | 2000-06-22 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Structurally Diverse Substrates by Type II 3-Hydroxyacyl-Coa Dehydrogenase (Hadh II) Amyloid-Beta Binding Alcohol Dehydrogenase (Abad)
J.Mol.Biol., 303, 2000
|
|
3PVF
 
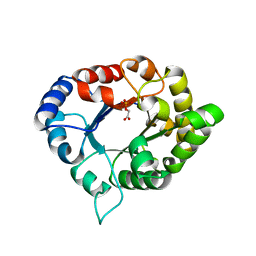 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
5FO5
 
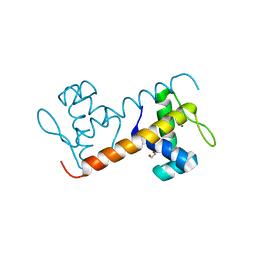 | | Structure of the DNA-binding domain of Escherichia coli methionine biosynthesis regulator MetR | | Descriptor: | 1,2-ETHANEDIOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR METR, MAGNESIUM ION | | Authors: | Punekar, A.S, Porter, J, Urbanowski, M.L, Stauffer, G.V, Carr, S.B, Phillips, S.E. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for DNA Recognition by the Transcription Regulator Metr.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2VFG
 
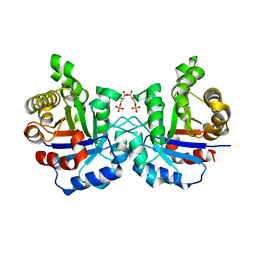 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase with 3-phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFD
 
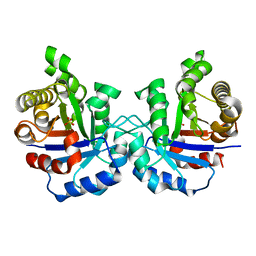 | | Crystal structure of the F96S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1R3O
 
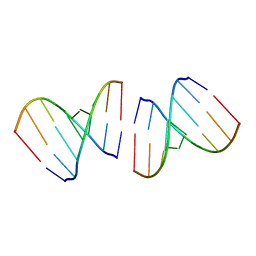 | | Crystal structure of the first RNA duplex in L-conformation at 1.9A resolution | | Descriptor: | L-RNA | | Authors: | Vallazza, M, Perbandt, M, Klussmann, S, Rypniewski, W, Erdmann, V.A, Betzel, C. | | Deposit date: | 2003-10-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First look at RNA in L-configuration.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2VFH
 
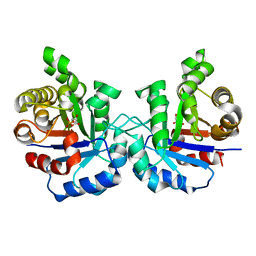 | | Crystal structure of the F96W mutant of Plasmodium falciparum triosephosphate isomerase complexed with 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4GUI
 
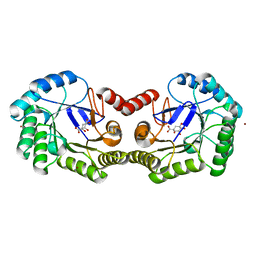 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4FN6
 
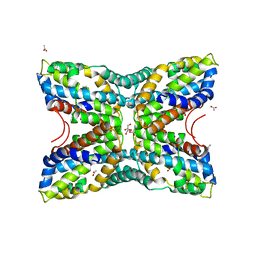 | | Structural Characterization of Thiaminase type II TenA from Staphylococcus aureus | | Descriptor: | ACETATE ION, GLYCEROL, thiaminase-2 | | Authors: | Begum, A, Drebes, J, Perbandt, M, Wrenger, C, Betzel, C. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Characterization of Thiaminase type II TenA from Staphylococcus aureus
TO BE PUBLISHED
|
|
3PWA
 
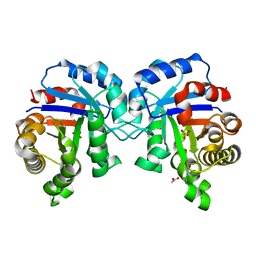 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
4GUJ
 
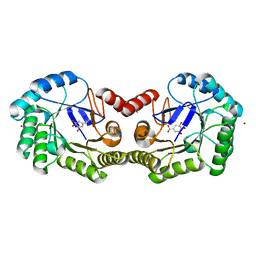 | | 1.50 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-dehydroquinate dehydratase, ZINC ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
2VFE
 
 | | Crystal structure of F96S mutant of Plasmodium falciparum triosephosphate isomerase with 3- phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCEROL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VFI
 
 | | Crystal structure of the Plasmodium falciparum triosephosphate isomerase in the loop closed state with 3-phosphoglycerate bound at the active site and interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1G8T
 
 | | SM ENDONUCLEASE FROM SERATIA MARCENSCENS AT 1.1 A RESOLUTION | | Descriptor: | MAGNESIUM ION, NUCLEASE SM2 ISOFORM, SULFATE ION | | Authors: | Lunin, V.V, Perbandt, M, Betzel, C.H, Mikhailov, A.M. | | Deposit date: | 2000-11-21 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structure of the Serratia marcescens endonuclease at 1.1 A resolution and the enzyme reaction mechanism.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3PY2
 
 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
2VFF
 
 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2V7R
 
 | | Crystal structure of a human tRNAGly microhelix at 1.2 Angstrom resolution | | Descriptor: | HUMAN TRNAGLY MICROHELIX | | Authors: | Foerster, C, Mankowska, M, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-08-01 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a Human Trnagly Microhelix at 1.2 A Resolution.
Biochem.Biophys.Res.Commun., 368, 2008
|
|
