2XMJ
 
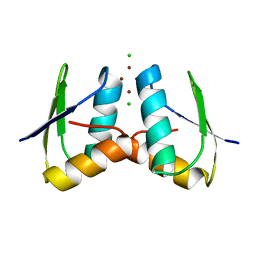 | | Visualising the Metal-binding Versatility of Copper Trafficking Sites: Atx1 side-to-side (aerobic) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SODIUM ION, ... | | Authors: | Badarau, A, Firbank, S.J, McCarthy, A.A, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Visualizing the Metal-Binding Versatility of Copper Trafficking Sites .
Biochemistry, 49, 2010
|
|
1P7V
 
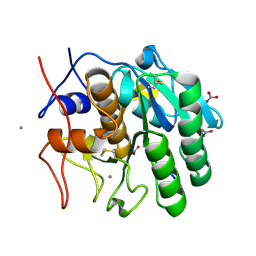 | | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Kaur, P, Chandra, V, Banumathi, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution
To be published
|
|
4BDA
 
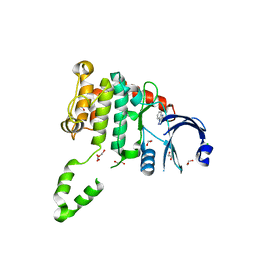 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydro[1,2,4]triazolo[1,5-a][3,1]benzimidazol-9-ium, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDK
 
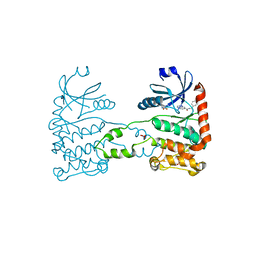 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHECKPOINT KINASE 2, N-[(4-methoxyphenyl)methyl]quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDG
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3-(PYRIDIN-3-YL)-1H-PYRAZOL-5-AMINE, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4A9U
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(1-BENZYLPIPERIDIN-4-YL)METHOXY]PHENYL}-1H-BENZIMIDAZOLE-6-CARBOXAMIDE, CHLORIDE ION, ... | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography
Bioorg.Med.Chem., 20, 2012
|
|
4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDI
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-acetyl-N-(5-methylpyridin-2-yl)piperidine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDC
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(furan-2-ylmethyl)quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
7P8K
 
 | | Crystal structure of in planta processed AvrRps4 in complex with the WRKY domain of RRS1 | | Descriptor: | Avirulence protein,Avirulence protein, Disease resistance protein RRS1, ZINC ION | | Authors: | Mukhi, N, Brown, H, Gorenkin, D, Ding, P, Bentham, A.R, Jones, J.D.G, Banfield, M.J. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4BDD
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDE
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 6-METHYLQUINAZOLIN-4-AMINE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
4BDF
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 5-METHYL-3-PHENYL-1H-PYRAZOLE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDJ
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 3-cyclopropyl-4-(furan-2-yl)-1H-pyrazolo[3,4-b]pyridine, CHECKPOINT KINASE 2, NITRATE ION | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDH
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYL-4-(THIOPHEN-2-YL)-1H-PYRAZOL-5-AMINE, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4A9T
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{[1-(4-CHLOROBENZYL)PIPERIDIN-4-YL]METHOXY}PHENYL)-1H-BENZIMIDAZOLE-5-CARBOXAMIDE, NITRATE ION, ... | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography.
Bioorg.Med.Chem., 20, 2012
|
|
4A9R
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(4-CHLOROPHENOXY)PHENYL]-1H-BENZIMIDAZOLE-6-CARBOXAMIDE, NITRATE ION, ... | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography
Bioorg.Med.Chem., 20, 2012
|
|
2MIW
 
 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | Authors: | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
4A9S
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 2-(4-(3-HYDROXYPHENOXY)PHENYL)-1H-BENZO[D]IMIDAZOLE-5-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography.
Bioorg.Med.Chem., 20, 2012
|
|
6Q76
 
 | |
2MIV
 
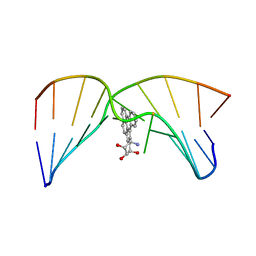 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
6RFU
 
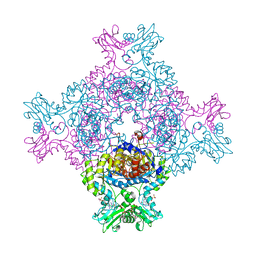 | | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of ATP and GMP as genuine co-factors | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase | | Authors: | Nass, K, Redecke, L, Perbandt, M, Yefanov, O, Gabdulkhakov, A, Duszenko, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co-factors.
Nat Commun, 11, 2020
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
