6WR4
 
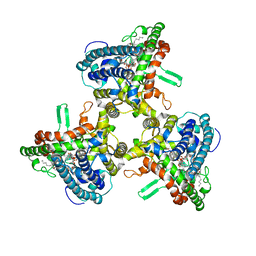 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
6U00
 
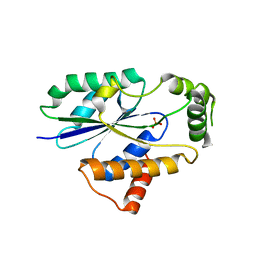 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZM
 
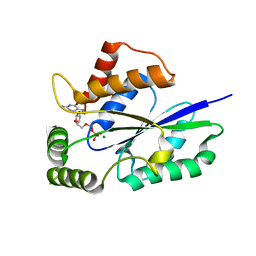 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6A1Q
 
 | | Crystal structures of disordered Z-type helices | | Descriptor: | DNA (5'-D(P*CP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*G)-3') | | Authors: | Karthik, S, Mandal, P.K, Thirugnanasambandam, A, Gautham, N. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of disordered Z-type helices.
Nucleosides Nucleotides Nucleic Acids, 38, 2019
|
|
5IYE
 
 | | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 resolution | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*CP*GP*GP*G)-3'), ZINC ION | | Authors: | Karthik, S, Thirugnanasambandam, A, Mandal, P.K, Gautham, N. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 angstrom resolution.
Nucleosides Nucleotides Nucleic Acids, 36, 2017
|
|
5IYG
 
 | |
5IYJ
 
 | |
5ZHP
 
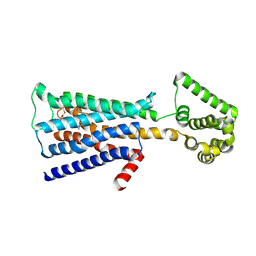 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WV7
 
 | |
6BK9
 
 | | Crystal Structure of Squid Arrestin | | Descriptor: | CHLORIDE ION, Visual arrestin | | Authors: | Eger, B.T, Bandyopadhyay, A, Yedidi, R.S, Ernst, O.P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00005579 Å) | | Cite: | A Novel Polar Core and Weakly Fixed C-Tail in Squid Arrestin Provide New Insight into Interaction with Rhodopsin.
J. Mol. Biol., 430, 2018
|
|
6BMM
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P21 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,5S)-hexane-2,5-diol, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
2LS5
 
 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
2LRN
 
 | | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp. | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-10 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thiol:disulfide interchange protein from Bacteroides sp.
To be Published
|
|
2LST
 
 | | Solution structure of a thioredoxin from Thermus thermophilus | | Descriptor: | Thioredoxin | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thioredoxin from Thermus thermophilus
To be Published
|
|
6E3A
 
 | | tRNA 2'-phosphotransferase | | Descriptor: | COENZYME A, Probable RNA 2'-phosphotransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2'-PO4recognition and ADP-ribosylation.
Nat Commun, 10, 2019
|
|
5YPP
 
 | | Crystal structure of IlvN.Val-1a | | Descriptor: | ACETATE ION, Acetolactate synthase isozyme 1 small subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
7JNO
 
 | |
8A28
 
 | |
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
7BML
 
 | |
6CMV
 
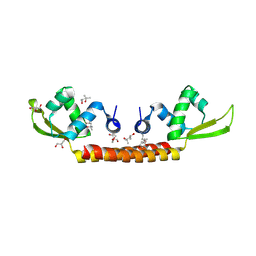 | | Crystal structure of Archaeal Biofilm Regulator (AbfR2) from Sulfolobus acidocaldarius | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Transcriptional regulator Lrs14-like protein | | Authors: | Essen, L.-O, Vogt, M.S, Banerjee, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an Lrs14-like archaeal biofilm regulator from Sulfolobus acidocaldarius.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8A9T
 
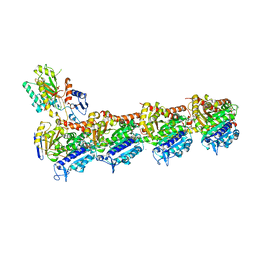 | | Tubulin-[1,2]oxazoloisoindole-1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
8A9Z
 
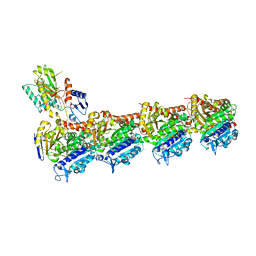 | | Tubulin-[1,2]oxazoloisoindole-2e complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-[(3,5-dimethoxyphenyl)methyl]pyrrolo[3,4-g][1,2]benzoxazole, CALCIUM ION, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
6BMN
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P63 | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ZINC ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6BML
 
 | | Structure of human DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
