6R7U
 
 | | Selenomethionine variant of Tannerella forsythia promirolysin mutant E225A | | Descriptor: | BORIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
6R7V
 
 | | Tannerella forsythia promirolysin mutant E225A | | Descriptor: | CALCIUM ION, GLYCEROL, Mirolysin, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
6R7W
 
 | | Tannerella forsythia mature mirolysin in complex with a cleaved peptide. | | Descriptor: | CALCIUM ION, CITRIC ACID, ETHANOL, ... | | Authors: | Rodriguez-Banqueri, A, Guevara, T, Ksiazek, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2019-03-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based mechanism of cysteine-switch latency and of catalysis by pappalysin-family metallopeptidases.
Iucrj, 7, 2020
|
|
7NEN
 
 | |
5KZE
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus | | Descriptor: | GLYCEROL, N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
5KZD
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus with bound sialic acid alditol | | Descriptor: | (2~{S},4~{S},5~{R},6~{R},7~{S},8~{R})-5-acetamido-2,4,6,7,8,9-hexakis(oxidanyl)nonanoic acid, N-acetylneuraminate lyase | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
2YLA
 
 | | INHIBITION OF THE PNEUMOCOCCAL VIRULENCE FACTOR STRH AND MOLECULAR INSIGHTS INTO N-GLYCAN RECOGNITION AND HYDROLYSIS | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
6ZA2
 
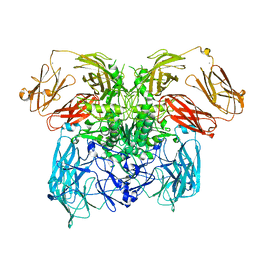 | | Crystal structure of dimeric latent PorU from Porphyromonas gingivalis | | Descriptor: | CALCIUM ION, Por secretion system protein porU | | Authors: | Gomis-Ruth, F.X, Goulas, T, Guevara, T, Rodriguez-Banqueri, A, Potempa, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Intermolecular latency regulates the essential C-terminal signal peptidase and sortase of the Porphyromonas gingivalis type-IX secretion system.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SKM
 
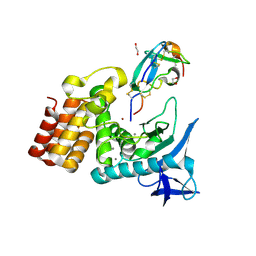 | | Complex between S. aureus aureolysin and wt IMPI. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
7SKL
 
 | | Complex between S. aureus aureolysin and IMPI mutant I57I | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, IMPI alpha, ... | | Authors: | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
8CD8
 
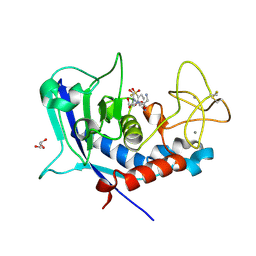 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
6Z8F
 
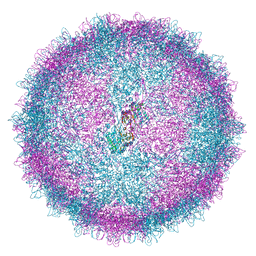 | | Human Picobirnavirus D45-CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6Z8D
 
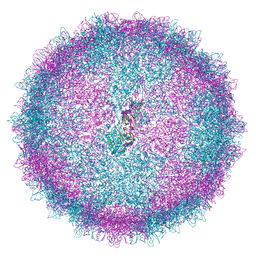 | | Human Picobirnavirus CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6Z8E
 
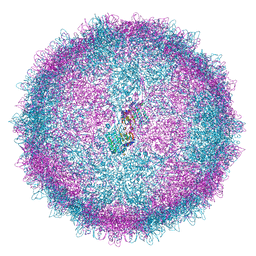 | | Human Picobirnavirus Ht-CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
8CDB
 
 | | Proulilysin E229A structure | | Descriptor: | CALCIUM ION, Ulilysin, ZINC ION | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
6I9A
 
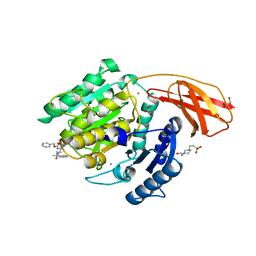 | | Porphyromonas gingivalis gingipain K (Kgp) in complex with inhibitor KYT-36 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Rofdriguez-Banqueri, A. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor.
Sci Rep, 9, 2019
|
|
8QB1
 
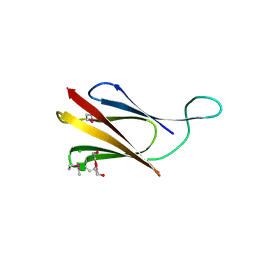 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system.
Open Biology, 14, 2024
|
|
3INO
 
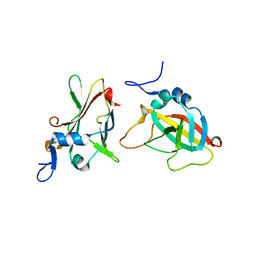 | | 1.95A Resolution Structure of Protective Antigen Domain 4 | | Descriptor: | Protective antigen PA-63 | | Authors: | Lovell, S, Williams, A.S, Anbanandam, A, El-Chami, R, Bann, J.G. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain 4 of the anthrax protective antigen maintains structure and binding to the host receptor CMG2 at low pH
Protein Sci., 18, 2009
|
|
8B2N
 
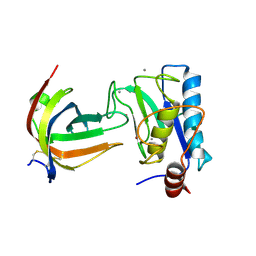 | | Potempin A (PotA) from Tannerella forsythia in complex with the catalytic domain of human MMP-12 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, Tannerella forsythia potempin A (PotA), ... | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
8XZ3
 
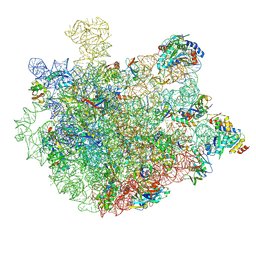 | |
8T3J
 
 | | Crystal structure of native exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Calil, F.A, Gismene, C, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
8T3I
 
 | | Crystal structure of mutant exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Gismene, C, Calil, F.A, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
7Z5F
 
 | | VP2-only capsid of MVM D263A mutant | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
7Z5E
 
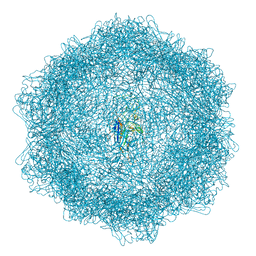 | | VP2-only capsid of MVM D263A mutant | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
7Z5D
 
 | | VP2-only capsid of wt MVM prototype strain p | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
