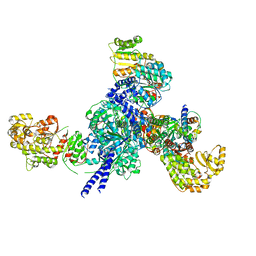
7S6B
 
 | |
7S6C
 
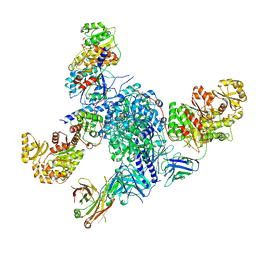 | | CryoEM structure of modular PKS holo-Lsd14 stalled at the condensation step and bound to antibody fragment 1B2, composite structure | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 4'-PHOSPHOPANTETHEINE, 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Bagde, S.R, Kim, C.-Y, Fromme, J.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modular polyketide synthase contains two reaction chambers that operate asynchronously.
Science, 374, 2021
|
|
7S6D
 
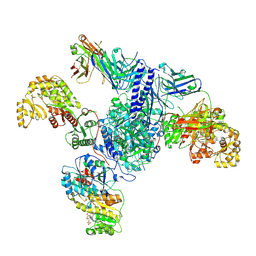 | | CryoEM structure of modular PKS holo-Lsd14 bound to antibody fragment 1B2, composite structure | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-deoxyerythronolide-B synthase EryA2, modules 3 and 4, ... | | Authors: | Bagde, S.R, Kim, C.-Y, Fromme, J.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modular polyketide synthase contains two reaction chambers that operate asynchronously.
Science, 374, 2021
|
|
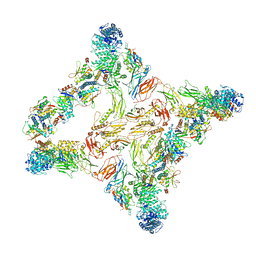
7U06
 
 | |
7U05
 
 | |
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | Descriptor: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
8GIA
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with TFMU-ADPr | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain.
Acs Chem.Biol., 18, 2023
|
|
8CTQ
 
 | | Crystal structure of engineered phospholipase D mutant superPLD 2-48 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
8CTP
 
 | | Crystal structure of engineered phospholipase D mutant superPLD 2-23 | | Descriptor: | PHOSPHATE ION, Phospholipase D | | Authors: | Tei, R, Bagde, S.R, Fromme, J.C, Baskin, J.M. | | Deposit date: | 2022-05-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based directed evolution of a membrane editor in mammalian cells.
Nat.Chem., 15, 2023
|
|
7BVZ
 
 | | Crystal structure of MreB5 of Spiroplasma citri bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell shape determining protein MreB, MAGNESIUM ION, ... | | Authors: | Pande, V, Bagde, S.R, Gayathri, P. | | Deposit date: | 2020-04-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MreB5 Is a Determinant of Rod-to-Helical Transition in the Cell-Wall-less Bacterium Spiroplasma.
Curr.Biol., 30, 2020
|
|
