7FSN
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z254580486 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSL
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z730649594 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FT0
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z56767614 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSX
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1328078283 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FT5
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with POB0093 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methoxyphenyl)oxane-4-carboxylic acid, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSO
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z198195770 | | Descriptor: | 1,2-ETHANEDIOL, D-GLUTAMIC ACID, GLYCINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSQ
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1429867185 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSS
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z839706072 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromanylpyrazol-1-yl)-~{N}-cyclopropyl-~{N}-methyl-ethanamide, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSZ
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z3006151474 | | Descriptor: | (5S)-5-(difluoromethoxy)pyridin-2(5H)-one, 1,2-ETHANEDIOL, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FT4
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z48847594 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FT3
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z291279160 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(trifluoromethyloxy)phenyl]thiourea, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
3P1A
 
 | | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Chaikuad, A, Eswaran, J, Fedorov, O, Cooper, C.D.O, Kroeler, T, Vollmar, M, Krojer, T, Berridge, G, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1)
To be Published
|
|
3PEH
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Endoplasmin homolog, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, Cossar, D, MacKenzie, F, Vedadi, M, Senisterra, G, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative
To be Published
|
|
3PFV
 
 | | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide | | Descriptor: | 1,2-ETHANEDIOL, 11-meric peptide from Epidermal growth factor receptor, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Guo, K, Cooper, C.D.O, Ayinampudi, V, Krojer, T, Muniz, J.R.C, Vollmar, M, Canning, P, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide
To be Published
|
|
3EB0
 
 | | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804 | | Descriptor: | 3-({[(3S)-3,4-dihydroxybutyl]oxy}amino)-1H,2'H-2,3'-biindol-2'-one, GLYCEROL, Putative uncharacterized protein | | Authors: | Wernimont, A.K, Fedorov, O, Lam, A, Ali, A, Zhao, Y, Lew, J, Wasney, G, Vedadi, M, Kozieradzki, I, Schapira, M, Bochkarev, A, Wilstrom, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of cgd4_240 from cryptosporidium Parvum in complex with indirubin E804
TO BE PUBLISHED
|
|
6RCG
 
 | | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, ~{N}-[[6,7-bis(fluoranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(3-fluorophenyl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Roush, W.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Casein kinase 1 delta (CK1 delta) complexed with SR3029 inhibitor
To Be Published
|
|
2JKU
 
 | | Crystal structure of the N-terminal region of the biotin acceptor domain of human propionyl-CoA carboxylase | | Descriptor: | PROPIONYL-COA CARBOXYLASE ALPHA CHAIN, MITOCHONDRIAL, TETRAETHYLENE GLYCOL | | Authors: | Healy, S, Yue, W.W, Kochan, G, Pilka, E.S, Murray, J.W, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Wikstrom, M, Edwards, A, Bountra, C, Gravel, R.A, Oppermann, U. | | Deposit date: | 2008-08-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment.
Biochemistry, 49, 2010
|
|
3L6Q
 
 | | Crystal structure of the N-terminal domain of HSP70 from Cryptosporidium parvum (CGD2_20) | | Descriptor: | Heat shock 70 (HSP70) protein, MAGNESIUM ION, SULFATE ION | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Lew, J, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of the N-terminal domain of HSP70 from Cryptosporidium parvum (CGD2_20)
To be Published
|
|
6ROG
 
 | | Crystal Structure of the KELCH domain of human KEAP1 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, SODIUM ION | | Authors: | Sethi, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bullock, A.N, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of the KELCH domain of human KEAP1
To Be Published
|
|
3LS8
 
 | | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol | | Descriptor: | 3-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Kraulis, P, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Van den Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
To be Published
|
|
3FI8
 
 | | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Pizarro, J.C, Artz, J.D, Amaya, M.F, Xiao, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020
TO BE PUBLISHED
|
|
3FK2
 
 | | Crystal structure of the RhoGAP domain of human glucocorticoid receptor DNA-binding factor 1 | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-15 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the RhoGAP domain of human glucocorticoid receptor DNA-binding factor 1
To be Published
|
|
3NR3
 
 | | Crystal Structure of Human Peripheral Myelin Protein 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Ugochukwu, E, Pilka, E, Phillips, C, Yue, W.W, Krojer, T, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peripheral Myelin Protein 2
To be Published
|
|
3CP6
 
 | | Crystal structure of human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor | | Descriptor: | (4aS,7aR)-octahydro-1H-cyclopenta[b]pyridine-6,6-diylbis(phosphonic acid), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, von Delft, F, Barnett, B.L, Ebetino, F.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor.
To be Published
|
|
6EL8
 
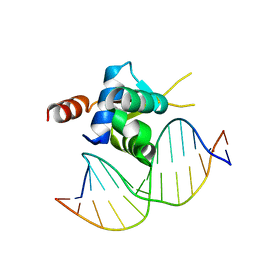 | | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*GP*CP*GP*TP*CP*TP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*AP*GP*AP*CP*GP*CP*CP*AP*CP*C)-3'), Forkhead box protein N1 | | Authors: | Newman, J.A, Aitkenhead, H.A, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA
To be published
|
|
