1F8E
 
 | | Native Influenza Neuraminidase in Complex with 4,9-diamino-2-deoxy-2,3-dehydro-N-acetyl-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,9-AMINO-2,4-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1FT7
 
 | | AAP COMPLEXED WITH L-LEUCINEPHOSPHONIC ACID | | Descriptor: | BACTERIAL LEUCYL AMINOPEPTIDASE, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Stamper, C, Bennett, B, Holz, R, Petsko, G, Ringe, D. | | Deposit date: | 2000-09-11 | | Release date: | 2000-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of the aminopeptidase from Aeromonas proteolytica by L-leucinephosphonic acid. Spectroscopic and crystallographic characterization of the transition state of peptide hydrolysis.
Biochemistry, 40, 2001
|
|
1FFO
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH SYNTHETIC PEPTIDE GP33 (C9M/K1A) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition.
J.Immunol., 169, 2002
|
|
1FG4
 
 | | STRUCTURE OF TRYPAREDOXIN II | | Descriptor: | TRYPAREDOXIN II | | Authors: | Hofmann, B, Budde, H, Bruns, K, Guerrero, S.A, Kalisz, H.M, Menge, U, Montemartini, M, Nogoceke, E, Steinert, P, Wissing, J.B, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-07-28 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
1F9S
 
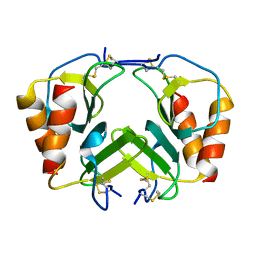 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 2 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1FFN
 
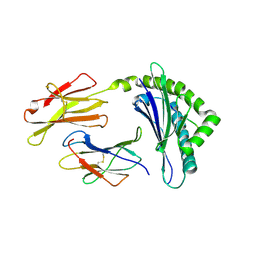 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33(C9M) | | Descriptor: | BETA-2 MICROGLOBULIN, BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
6DNL
 
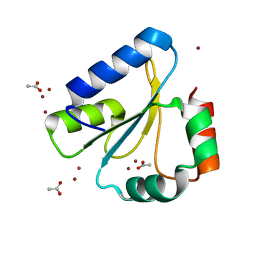 | | Crystal Structure of Neisseria meningitidis DsbD c-terminal domain in the reduced form | | Descriptor: | ACETATE ION, Thiol:disulfide interchange protein DsbD, ZINC ION | | Authors: | Smith, R.P, Heras, B, Paxman, J.J. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the disulfide reductase mechanism of DsbD, an essential enzyme for neisserial pathogens.
J. Biol. Chem., 293, 2018
|
|
1FSB
 
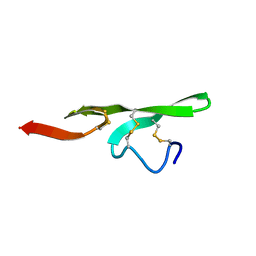 | | STRUCTURE OF THE EGF DOMAIN OF P-SELECTIN, NMR, 19 STRUCTURES | | Descriptor: | P-SELECTIN | | Authors: | Freedman, S.J, Sanford, D.G, Bachovchin, W.W, Furie, B.C, Baleja, J.D, Furie, B. | | Deposit date: | 1996-03-25 | | Release date: | 1997-04-01 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the epidermal growth factor domain of P-selectin.
Biochemistry, 35, 1996
|
|
1FFX
 
 | | TUBULIN:STATHMIN-LIKE DOMAIN COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PROTEIN (STATHMIN-LIKE DOMAIN OF RB3), ... | | Authors: | Gigant, B, Martin-Barbey, C, Knossow, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | The 4 A X-ray structure of a tubulin:stathmin-like domain complex.
Cell(Cambridge,Mass.), 102, 2000
|
|
4MX6
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella oneidensis (SO_3134), target EFI-510275, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP-type C4-dicarboxylate:H+ symport system substrate-binding component DctP | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-25 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1FG8
 
 | | STRUCTURAL IMPLICATIONS OF DRUG RESISTANT MUTANTS OF HIV-1 PROTEASE: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE MUTANT PROTEASE/SUBSTRATE ANALOG COMPLEXES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2000-07-25 | | Release date: | 2001-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes.
Proteins, 43, 2001
|
|
4N6D
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1FPU
 
 | | CRYSTAL STRUCTURE OF ABL KINASE DOMAIN IN COMPLEX WITH A SMALL MOLECULE INHIBITOR | | Descriptor: | N-[4-METHYL-3-[[4-(3-PYRIDINYL)-2-PYRIMIDINYL]AMINO]PHENYL]-3-PYRIDINECARBOXAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL | | Authors: | Schindler, T, Bornmann, W, Pellicena, P, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism for STI-571 inhibition of abelson tyrosine kinase.
Science, 289, 2000
|
|
4N4U
 
 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NAP
 
 | | Crystal structure of a trap periplasmic solute binding protein from Desulfovibrio alaskensis G20 (DDE_0634), target EFI-510102, with bound d-tryptophan | | Descriptor: | D-TRYPTOPHAN, Extracellular solute-binding protein, family 7 | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8G
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0660), Target EFI-501075, with bound D-alanine-D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NGU
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio alaskensis G20 (Dde_1548), Target EFI-510103, with bound D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-02 | | Release date: | 2013-12-04 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NHB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio desulfuricans (Ddes_1525), Target EFI-510107, with bound sn-glycerol-3-phosphate | | Descriptor: | IODIDE ION, SN-GLYCEROL-3-PHOSPHATE, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-04 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NF0
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOMONAS AERUGINOSA PAO1 (PA4616), TARGET EFI-510182, WITH BOUND L-Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable c4-dicarboxylate-binding protein, SULFATE ION | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NG7
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Citrobacter koseri (CKO_04899), Target EFI-510094, apo, open structure | | Descriptor: | TRAP periplasmic solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6EPM
 
 | | Ras guanine nucleotide exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) and fragment screening hit F1 | | Descriptor: | (1-phenyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)methanamine, GLYCEROL, GTPase KRas, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EPP
 
 | | RAS GUANINE EXCHANGE FACTOR SOS1 (REM-CDC25) IN COMPLEX WITH KRAS(G12C) AND FRAGMENT SCREENING HIT F4 | | Descriptor: | GLYCEROL, GTPase KRas, Son of sevenless homolog 1, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EPN
 
 | | Ras guanine exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) and fragment screening hit F2 | | Descriptor: | 1-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-2-oxidanyl-ethanone, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6E8R
 
 | |
4F7T
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PB1 (498-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
