4K1Z
 
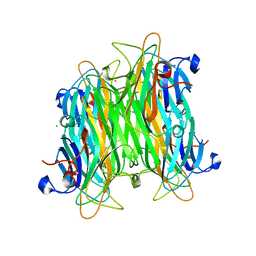 | | Crystal structure of Canavalia boliviana lectin in complex with Man1-4Man-OMe | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bezerra, G.A, Viertlmayr, R, Moura, T.R, Delatorre, P, Rocha, B.A.M, Cavada, B.S, Gruber, K. | | Deposit date: | 2013-04-07 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies of an Anti-Inflammatory Lectin from Canavalia boliviana Seeds in Complex with Dimannosides.
Plos One, 9, 2014
|
|
8CQF
 
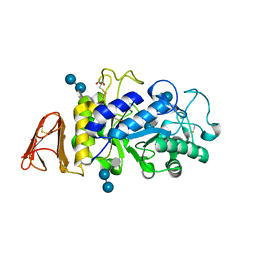 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis Complexed with Rearranged Acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skagseth, S, Griese, J.J, Lund, B.A, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
8CQG
 
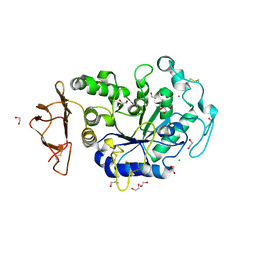 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis | | Descriptor: | 1,2-ETHANEDIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Skagseth, S, Lund, B.A, Griese, J.J, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
4KBL
 
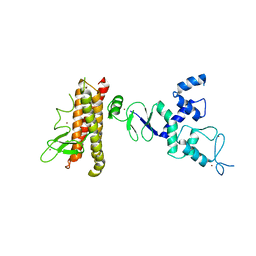 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
1GWI
 
 | | The 1.92 A structure of Streptomyces coelicolor A3(2) CYP154C1: A new monooxygenase that functionalizes macrolide ring systems | | Descriptor: | CYTOCHROME P450 154C1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Podust, L.M, Kim, Y, Arase, M, Neely, B.A, Beck, B.J, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2002-03-15 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.92 A Structure of Streptomyces Coelicolor A3(2) Cyp154C1: A New Monooxygenase that Functionalizes Macrolide Ring Systems
J.Biol.Chem., 278, 2003
|
|
1I4A
 
 | | CRYSTAL STRUCTURE OF PHOSPHORYLATION-MIMICKING MUTANT T6D OF ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION, SULFATE ION | | Authors: | Kaetzel, M.A, Mo, Y.D, Mealy, T.R, Campos, B, Bergsma-Schutter, W, Brisson, A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2001-02-20 | | Release date: | 2001-04-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation mutants elucidate the mechanism of annexin IV-mediated membrane aggregation.
Biochemistry, 40, 2001
|
|
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF9
 
 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8E84
 
 | | Human PCNA mutant- C148S | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Magrino, J, Page, B, Gaubitz, C, Kelch, B.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A thermosensitive PCNA allele underlies an ataxia-telangiectasia-like disorder.
J.Biol.Chem., 299, 2023
|
|
7ZVO
 
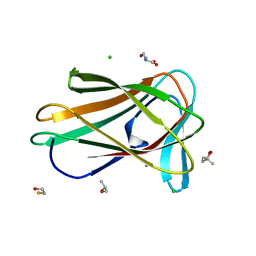 | | Structure of CBM BT0996-C from Bacteroides thetaiotaomicron | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALANINE, BETA-MERCAPTOETHANOL, ... | | Authors: | Trovao, F, Pinheiro, B.A, Correia, V.G, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2022-05-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a Bacteroides thetaiotaomicron carbohydrate-binding module provides new insight into the recognition of complex pectic polysaccharides by the human microbiome.
J Struct Biol X, 7, 2023
|
|
2ADX
 
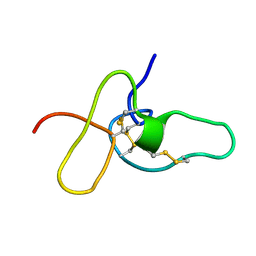 | | FIFTH EGF-LIKE DOMAIN OF THROMBOMODULIN (TMEGF5), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | THROMBOMODULIN | | Authors: | Sampoli-Benitez, B.A, Hunter, M.J, Meininger, D.P, Komives, E.A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-12-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth EGF-like domain of thrombomodulin: An EGF-like domain with a novel disulfide-bonding pattern.
J.Mol.Biol., 273, 1997
|
|
2XHB
 
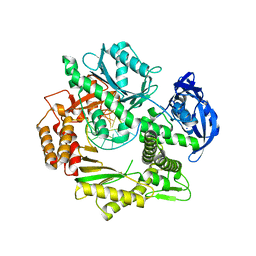 | | Crystal structure of DNA polymerase from Thermococcus gorgonarius in complex with hypoxanthine-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', DNA POLYMERASE, HYPOXANTHINE-CONTAINING DNA, ... | | Authors: | Killelea, T, Ghosh, S, Tan, S.S, Heslop, P, Firbank, S.J, Kool, E.T, Connolly, B.A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Probing the Interaction of Archaeal DNA Polymerases with Deaminated Bases Using X-Ray Crystallography and Non-Hydrogen Bonding Isosteric Base Analogues.
Biochemistry, 49, 2010
|
|
4KC9
 
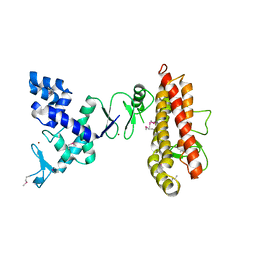 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
8E7I
 
 | |
8E7H
 
 | |
8E7E
 
 | |
8E7J
 
 | |
8E7D
 
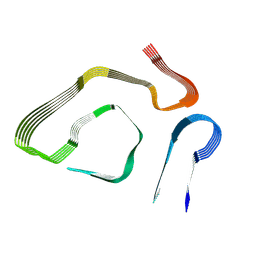 | |
1HMV
 
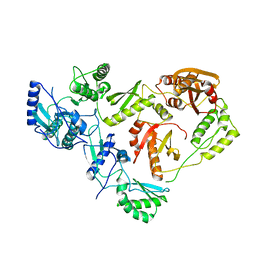 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
4IUM
 
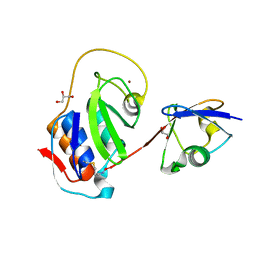 | | Equine arteritis virus papain-like protease 2 (PLP2) covalently bound to ubiquitin | | Descriptor: | GLYCEROL, Ubiquitin, ZINC ION, ... | | Authors: | Bailey-Elkin, B.A, James, T.W, Mark, B.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8B5P
 
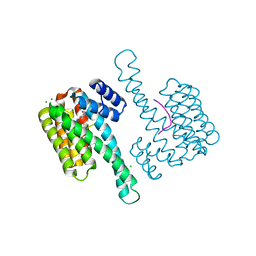 | |
8BI7
 
 | | Binary structure of 14-3-3s and PKR phosphopeptide | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, PKR phosphopeptide | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Dual-Covalent Molecular Locking of the 14-3-3/ERR gamma Protein-Protein Interaction as a Molecular Glue Drug Discovery Approach.
J.Am.Chem.Soc., 145, 2023
|
|
8B2I
 
 | |
8B4Q
 
 | | Ternary structure of 14-3-3s, ERRg phosphopeptide and dual-reactive compound 5 | | Descriptor: | 14-3-3 protein sigma, 3-fluoranyl-4-methanoyl-~{N}-methyl-~{N}-(2-sulfanylethyl)benzamide, CHLORIDE ION, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2022-09-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Dual-Covalent Molecular Locking of the 14-3-3/ERR gamma Protein-Protein Interaction as a Molecular Glue Drug Discovery Approach.
J.Am.Chem.Soc., 145, 2023
|
|
