1DVF
 
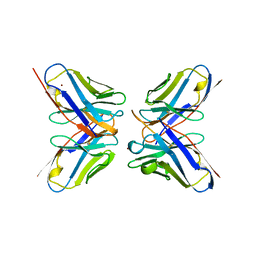 | | IDIOTOPIC ANTIBODY D1.3 FV FRAGMENT-ANTIIDIOTOPIC ANTIBODY E5.2 FV FRAGMENT COMPLEX | | Descriptor: | FV D1.3, FV E5.2, ZINC ION | | Authors: | Braden, B.C, Fields, B.A, Ysern, X, Dall'Acqua, W, Goldbaum, F.A, Poljak, R.J, Mariuzza, R.A. | | Deposit date: | 1996-04-13 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an Fv-Fv idiotope-anti-idiotope complex at 1.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
7R1K
 
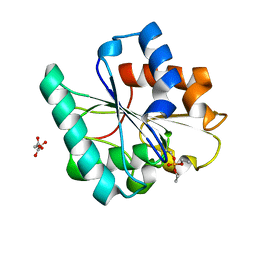 | | Phosphorylated Bacillus pumilus Lipase A | | Descriptor: | DIETHYL PHOSPHONATE, Lipase, OXALOACETATE ION | | Authors: | Lund, B.A. | | Deposit date: | 2022-02-03 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of a Cold-Adapted Bacterial Lipase
Biochemistry, 61, 2022
|
|
7R25
 
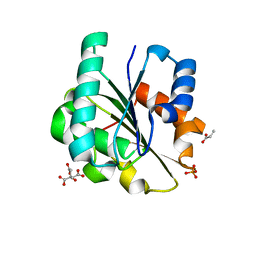 | | Bacillus pumilus Lipase A | | Descriptor: | CITRIC ACID, Lipase, PHOSPHATE ION, ... | | Authors: | Lund, B.A. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Structure and Mechanism of a Cold-Adapted Bacterial Lipase
Biochemistry, 61, 2022
|
|
2C54
 
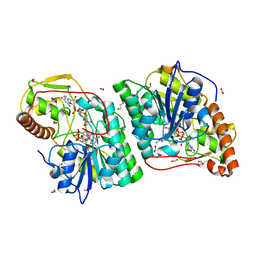 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana),k178r, with gdp-beta-l-gulose and gdp-4-keto-beta-l-gulose bound in active site. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of GDP-mannose-3',5'-epimerase: an enzyme which performs three chemical reactions at the same active site.
J. Am. Chem. Soc., 127, 2005
|
|
1EDN
 
 | | HUMAN ENDOTHELIN-1 | | Descriptor: | ENDOTHELIN-1 | | Authors: | Wallace, B.A, Janes, R.W. | | Deposit date: | 1994-09-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The crystal structure of human endothelin.
Nat.Struct.Biol., 1, 1994
|
|
1EJF
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CO-CHAPERONE P23 | | Descriptor: | Prostaglandin E synthase 3, SULFATE ION | | Authors: | Weaver, A.J, Sullivan, W.P, Felts, S.J, Owen, B.A.L, Toft, D.O. | | Deposit date: | 2000-03-02 | | Release date: | 2000-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
J.Biol.Chem., 275, 2000
|
|
1EEM
 
 | | GLUTATHIONE TRANSFERASE FROM HOMO SAPIENS | | Descriptor: | GLUTATHIONE, GLUTATHIONE-S-TRANSFERASE, SULFATE ION | | Authors: | Board, P, Coggan, M, Chelvanayagam, G, Easteal, S, Jermiin, L.S, Schulte, G.K, Danley, D.E, Hoth, L.R, Griffor, M.C, Kamath, A.V, Rosner, M.H, Chrunyk, B.A, Perregaux, D.E, Gabel, C.A, Geoghegan, K.F, Pandit, J. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and crystal structure of the Omega class glutathione transferases.
J.Biol.Chem., 275, 2000
|
|
1EKW
 
 | | NMR STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*TP*CP*GP*CP*AP*GP*C)-3') | | Authors: | Thiviyanathan, V, Luxon, B.A, Leontis, N.B, Donne, D, Gorenstein, D.G. | | Deposit date: | 2000-03-09 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY.
J.Biomol.NMR, 14, 1999
|
|
1EPP
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EON
 
 | | ECORV BOUND TO 3'-S-PHOSPHOROTHIOLATE DNA AND CA2+ | | Descriptor: | ACETIC ACID, CHLORIDE ION, DNA (5'-D(*AP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
7R4B
 
 | | The Bacillus pumilus chorismate mutase | | Descriptor: | 1,2-ETHANEDIOL, Chorismate mutase AroH | | Authors: | Lund, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Bacillus pumilus chorismate mutase
To Be Published
|
|
3URE
 
 | |
1EO3
 
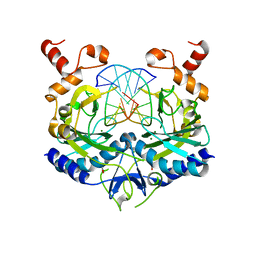 | | INHIBITION OF ECORV ENDONUCLEASE BY DEOXYRIBO-3'-S-PHOSPHOROTHIOLATES: A HIGH RESOLUTION X-RAY CRYSTALLOGRAPHIC STUDY | | Descriptor: | ACETIC ACID, DNA (5'-D(*CP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
7RC0
 
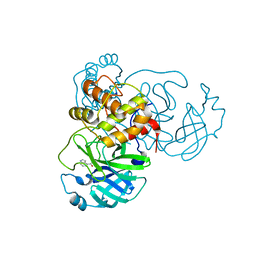 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-091-20 | | Descriptor: | 3C-like proteinase, 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate, SODIUM ION | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RC1
 
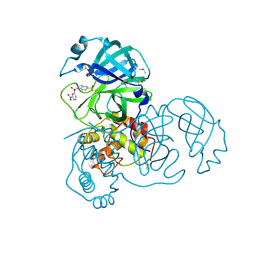 | | X-ray Structure of SARS-CoV main protease covalently modified by compound GRL-0686 | | Descriptor: | 3C-like proteinase, 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate, DIMETHYL SULFOXIDE | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
3URC
 
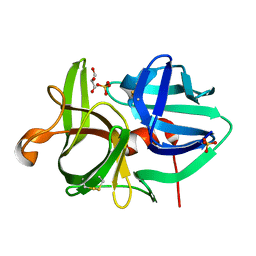 | | T181G mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7RBZ
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-017-20 | | Descriptor: | 3C-like proteinase, 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | | Authors: | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
1EO4
 
 | | ECORV BOUND TO MN2+ AND COGNATE DNA CONTAINING A 3'S SUBSTITION AT THE CLEAVAGE SITE | | Descriptor: | ACETIC ACID, DNA (5'-D(*CP*AP*AP*GP*AP*(TSP)P*AP*TP*CP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Horton, N.C, Connolly, B.A, Perona, J.J. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of EcoRV Endonuclease by Deoxyribo-3'-S-phosphorothiolates: A High-Resolution X-ray Crystallographic Study
J.Am.Chem.Soc., 122, 2000
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YOV
 
 | |
3URD
 
 | | T181A mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2C5A
 
 | | GDP-mannose-3', 5' -epimerase (Arabidopsis thaliana),Y174F, with GDP-beta-L-galactose bound in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
2BSL
 
 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE A, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-05-23 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
7PZW
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (4S)-N-((3R,5R)-1-(cyclopropanecarbonyl)-5-((5-(phenylethynyl)thiophene-2-carboxamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
3TEW
 
 | |
