6PG2
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product H5 (morpholine 8) | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PBI
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine 8 | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PGJ
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product A5 (Morpholine carboxylic acid 7) | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, (3S)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG1
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product F1 (methylpiperazinone 6) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PDH
 
 | | Crystal Structure of EcDsbA in a complex with purified pyrazole 9 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PD7
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine carboxylic acid 7 | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-18 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
2KQ2
 
 | | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A | | Descriptor: | Ribonuclease H-related protein | | Authors: | Mills, J.L, Eletsky, A, Hua, J, Belote, R.L, Buchwald, W.A, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apo form of a ribonuclease H domain of protein DSY1790 from Desulfitobacterium hafniense, Northeast Structural Genomics target DhR1A
To be Published
|
|
2KVU
 
 | | Solution NMR Structure of SAP domain of MKL/myocardin-like protein 1 from H.sapiens, Northeast Structural Genomics Consortium Target Target HR4547E | | Descriptor: | MKL/myocardin-like protein 1 | | Authors: | Liu, G, Xiao, R, Janjua, J, Acton, T.B, Mao, B, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-28 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4547E
To be Published
|
|
2KV7
 
 | | NMR solution structure of a soluble PrgI mutant from Salmonella Typhimurium | | Descriptor: | Protein prgI | | Authors: | Schmidt, H, Poyraz, O, Seidel, K, Delissen, F, Ader, C, Tenenboim, H, Goosmann, C, Laube, B, Thuenemann, A.F, Zychlinski, A, Baldus, M, Lange, A, Griesinger, C, Kolbe, M. | | Deposit date: | 2010-03-09 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein refolding is required for assembly of the type three secretion needle.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KUB
 
 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
2KWG
 
 | | Solution structure of a fully modified 2'-F/2'-OMe siRNA construct | | Descriptor: | 5'-R(*(GF2)P*(OMG)P*(GF2)P*(OMU)P*(AF2)P*(A2M)P*(AF2)P*(OMU)P*(AF2)P*(OMC)P*(AF2)P*(OMU)P*(UFT)P*(OMC)P*(UFT)P*(OMU)P*(CFZ)P*(A2M)P*(UFT)P*(OMU)P*(UFT))-3', 5'-R(P*(A2M)P*(UFT)P*(OMG)P*(AF2)P*(A2M)P*(GF2)P*(A2M)P*(AF2)P*(OMU)P*(GF2)P*(OMU)P*(AF2)P*(OMU)P*(UFT)P*(OMU)P*(AF2)P*(OMC)P*(CFZ)P*(OMC)P*(UFT)P*(OMU))-3' | | Authors: | Podbevsek, P, Bhat, B, Plavec, J. | | Deposit date: | 2010-04-09 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a fully alternately 2'-F/2'-OMe modified 42-nt dimeric siRNA construct.
Nucleic Acids Res., 38, 2010
|
|
2KYK
 
 | | The sandwich region between two LMP2A PY motif regulates the interaction between AIP4WW2domain and PY motif | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Seo, M, Park, S, Seok, S, Kim, J, Cha, M, Lee, B. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The sandwich region between two LMP2A PY motif regulates the interaction between AIP4WW2domain and PY motif
To be Published
|
|
2KYR
 
 | | Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12. Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544 | | Descriptor: | Fructose-like phosphotransferase enzyme IIB component 1 | | Authors: | Wu, B, Yee, A, Gutmanas, A, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Enzyme IIB subunit of PTS system from Escherichia coli K12, Northeast Structural Genomics Consortium target ER315/Ontario Center for Structural Proteomics target ec0544
To be Published
|
|
2KVY
 
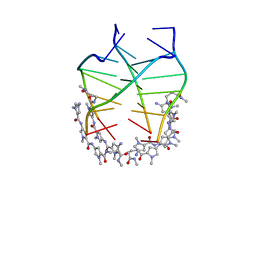 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
2KYT
 
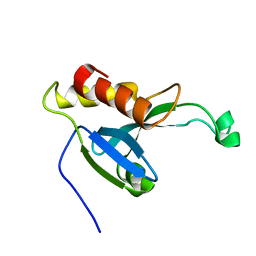 | | Solution structure of the H-REV107 N-terminal domain | | Descriptor: | Group XVI phospholipase A2 | | Authors: | Ren, X, Xia, B. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal catalytic domain of human H-REV107--a novel circular permutated NlpC/P60 domain
Febs Lett., 584, 2010
|
|
2KWB
 
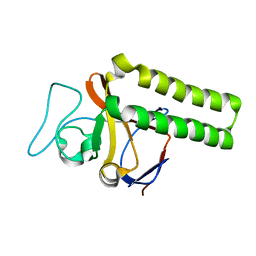 | | Minimal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from C.elegans, Northeast Structural Genomics Consortium Target WR73 | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Aramini, J.M, Rossi, P, Cort, J.R, Cooper, B, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Minimal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from C.elegans, Northeast Structural Genomics Consortium Target WR73
To be Published
|
|
2KD0
 
 | | NMR solution structure of O64736 protein from Arabidopsis thaliana. Northeast Structural Genomics Consortium MEGA Target AR3445A | | Descriptor: | LRR repeats and ubiquitin-like domain-containing protein At2g30105 | | Authors: | Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-31 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of O64736 protein from Arabidopsis thaliana
To be Published
|
|
2K4X
 
 | | Solution structure of 30S ribosomal protein S27A from Thermoplasma acidophilum | | Descriptor: | 30S ribosomal protein S27ae, ZINC ION | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 30S ribosomal protein S27A from Thermoplasma acidophilum/Northeast Structural Genomics Consortium Target TaT88/Ontario Center for Structural Proteomics target ta1093
To be Published
|
|
2KVS
 
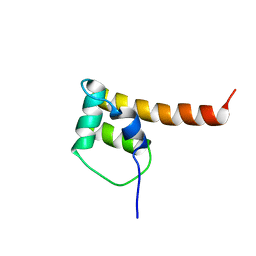 | | NMR Solution Structure of Q7A1E8 protein from Staphylococcus aureus: Northeast Structural Genomics Consortium target: ZR215 | | Descriptor: | Uncharacterized protein MW0776 | | Authors: | Swapna, G.V.T, Montelione, A.F, Wang, D, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Rost, B, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-27 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Q7A1E8 protein from Staphylococcus aureus: Northeast Structural Genomics Consortium target: ZR215
To be Published
|
|
2K5D
 
 | | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108]. | | Descriptor: | uncharacterized protein SAG0934 | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Foote, E.L, Jiang, M, Xiao, R, Sharma, S, Swapna, G.VT, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108].
To be Published
|
|
2KOB
 
 | | Solution NMR structure of CLOLEP_01837 (fragment 61-160) from Clostridium leptum. Northeast Structural Genomics Consortium Target QlR8A | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Lee, D, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of CLOLEP_01837 (fragment 61-160) from Clostridium leptum. Northeast Structural Genomics Consortium Target QlR8A
To be Published
|
|
2KRS
 
 | | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. | | Descriptor: | Probable enterotoxin | | Authors: | Ramelot, T.A, Cort, J.R, Maglaqui, M, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A.
To be Published
|
|
2KRK
 
 | | Solution NMR Structure of 26S protease regulatory subunit 8 from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3102A | | Descriptor: | 26S protease regulatory subunit 8 | | Authors: | Liu, G, Janjua, J, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of 26S protease regulatory subunit 8 from H.sapiens, Northeast Structural Genomics Consortium Target HR3102A
To be Published
|
|
2KWQ
 
 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
2KYY
 
 | | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea, Northeast Structural Genomics Consortium Target NeR70A | | Descriptor: | Possible ATP-dependent DNA helicase RecG-related protein | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Maglaqui, M, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea
To be Published
|
|
