6E4L
 
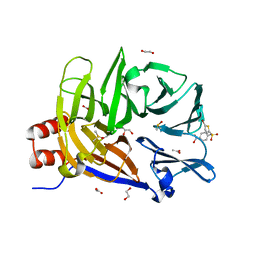 | | The structure of the N-terminal domain of human clathrin heavy chain 1 (nTD) in complex with ES9 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(4-nitrophenyl)thiophene-2-sulfonamide, ACETATE ION, ... | | Authors: | Dejonghe, W, Sharma, I, Denoo, B, Munck, S.D, Bulut, H, Mylle, E, Vasileva, M, Lu, Q, Savatin, D.V, Mishev, K, Nerinckx, W, Staes, A, Drozdzecki, A, Audenaert, D, Madder, A, Friml, J, Damme, D.V, Gevaert, K, Haucke, V, Savvides, S, Winne, J, Russinova, E. | | Deposit date: | 2018-07-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of endocytosis through chemical inhibition of clathrin heavy chain function.
Nat.Chem.Biol., 15, 2019
|
|
4MBS
 
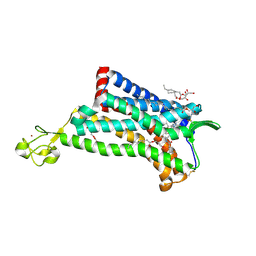 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
4ME5
 
 | |
4MG2
 
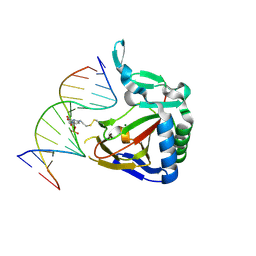 | | ALKBH2 F102A cross-linked to undamaged dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA-1, DNA-2, ... | | Authors: | Chen, B, Gan, J, Yang, C.G. | | Deposit date: | 2013-08-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex structures of ALKBH2 mutants cross-linked to
dsDNA reveal the conformational swing of β-hairpin
Sci China Chem, 57, 2014
|
|
6ERI
 
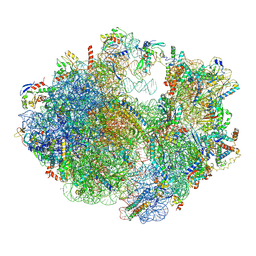 | | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10 alpha, ... | | Authors: | Perez Borema, A, Aibara, S, Paul, B, Tobiasson, V, Kimanius, D, Forsberg, B.O, Wallden, K, Lindahl, E, Amunts, A. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor.
Nat Plants, 4, 2018
|
|
4M9C
 
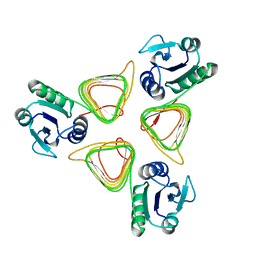 | | WeeI from Acinetobacter baumannii AYE | | Descriptor: | Bacterial transferase hexapeptide (Three repeats) family protein | | Authors: | Morrison, M.J, Imperiali, B. | | Deposit date: | 2013-08-14 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical analysis and structure determination of bacterial acetyltransferases responsible for the biosynthesis of UDP-N,N'-diacetylbacillosamine.
J.Biol.Chem., 288, 2013
|
|
6L3U
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the presence of calcium | | Descriptor: | CALCIUM ION, Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
4LTM
 
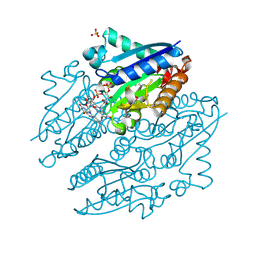 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
6E8A
 
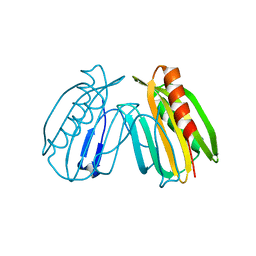 | | Crystal structure of DcrB from Salmonella enterica at 1.92 Angstroms resolution | | Descriptor: | DUF1795 domain-containing protein | | Authors: | Rasmussen, D.M, Soens, R.W, Bhattacharyya, B, May, J.F. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The structure of DcrB, a lipoprotein from Salmonella enterica, reveals flexibility in the N-terminal segment of the Mog1p/PsbP-like fold.
J. Struct. Biol., 204, 2018
|
|
4LUZ
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
6EBA
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Unoccupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-08-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6ENE
 
 | | OmpF orthologue from Enterobacter cloacae (OmpE35) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein (Porin), SULFATE ION, ... | | Authors: | van den Berg, B, Abellon-Ruiz, J, Basle, A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
4LZP
 
 | |
6EO6
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1H-indol-3-yl)acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA63A - TBA MODIFIED APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6L04
 
 | | Crystal structure of uPA_H99Y in complex with 31F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of uPA_H99Y in complex with 31F
To Be Published
|
|
4M1A
 
 | | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldella termitidis ATCC 33386 | | Descriptor: | Hypothetical protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldellatermitidis ATCC 33386
To be Published
|
|
4MI1
 
 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Microcin immunity protein MccF, SULFATE ION | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
6ES0
 
 | | Crystal structure of the kinase domain of human RIPK2 in complex with the activation loop targeting inhibitor CS-R35 | | Descriptor: | 2-[2-fluoranyl-4-[[2-fluoranyl-4-[2-(methylcarbamoyl)pyridin-4-yl]oxy-phenyl]carbamoylamino]phenyl]sulfanylethanoic acid, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pinkas, D.M, Bufton, J.C, Suebsuwong, C, Ray, S.S, Dai, B, Newman, J.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Degterev, A, Cuny, G.D, Bullock, A.N. | | Deposit date: | 2017-10-19 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Activation loop targeting strategy for design of receptor-interacting protein kinase 2 (RIPK2) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6E8J
 
 | | Crystal Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster, in Inward-facing And Occupied Conformation | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
4MN5
 
 | | Crystal structure of PAS domain of S. aureus YycG | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Shaikh, N, Hvorup, R, Winnen, B, Collins, B.M, King, G.F. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PAS domain of S. aureus YycG
To be Published
|
|
6L8S
 
 | | High resolution crystal structure of crustacean hemocyanin. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Masuda, T, Mikami, B, Baba, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Arch.Biochem.Biophys., 688, 2020
|
|
6EHD
 
 | |
4MM1
 
 | | GGGPS from Methanothermobacter thermautotrophicus | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE, TRIETHYLENE GLYCOL | | Authors: | Rajendran, C, Peterhoff, D, Beer, B, Kumpula, E.P, Kapetaniou, E, Guldan, H, Wierenga, R.K, Sterner, R, Babinger, P. | | Deposit date: | 2013-09-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8004 Å) | | Cite: | A comprehensive analysis of the geranylgeranylglyceryl phosphate synthase enzyme family identifies novel members and reveals mechanisms of substrate specificity and quaternary structure organization.
Mol.Microbiol., 92, 2014
|
|
4MN6
 
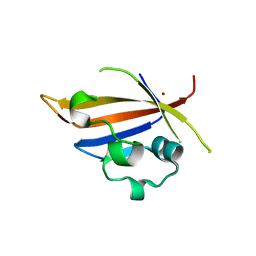 | | Crystal structure of truncated PAS domain from S. aureus YycG | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Shaikh, N, Hvorup, R, Winnen, B, Collins, B.M, King, G.F. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PAS domain of S. aureus YycG
To be Published
|
|
4MJX
 
 | |
