4W5A
 
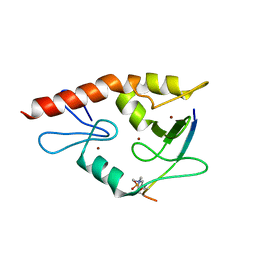 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V3B
 
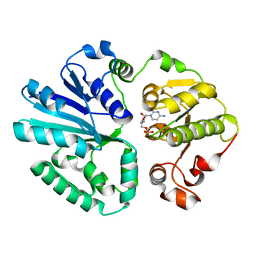 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4V4X
 
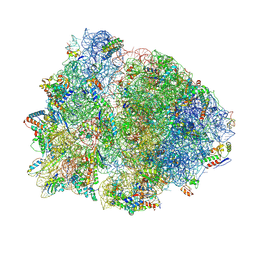 | | Crystal structure of the 70S Thermus thermophilus ribosome showing how the 16S 3'-end mimicks mRNA E and P codons. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
6XGT
 
 | |
6XWT
 
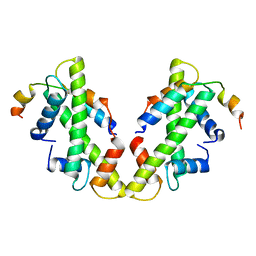 | | drosophila melanogaster CENP-A/H4 bound to N-terminal CAL1 fragment | | Descriptor: | Chromosome alignment defect 1, Histone H3-like centromeric protein cid, Histone H4 | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
6XYO
 
 | | Multiple system atrophy Type I alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6XSE
 
 | |
6XSH
 
 | |
6XQB
 
 | | SARS-CoV-2 RdRp/RNA complex | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
4V2I
 
 | | Biochemical characterization and structural analysis of a new cold- active and salt tolerant esterase from the marine bacterium Thalassospira sp | | Descriptor: | ESTERASE/LIPASE, MAGNESIUM ION | | Authors: | Santi, C.D, Leiros, H.-K.S, Scala, A.D, Pascale, D.D, Altermark, B, Willassen, N.-P. | | Deposit date: | 2014-10-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
Extremophiles, 20, 2016
|
|
4FS8
 
 | | The structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation | | Descriptor: | Arsenic methyltransferase, CALCIUM ION | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
4FSK
 
 | | Urate oxidase-azide complex in anaerobic conditions | | Descriptor: | AZIDE ION, SODIUM ION, Uricase | | Authors: | Gabison, L, Colloc'h, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2012-06-27 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Azide and Cyanide Have Different Inhibition Modes of Urate Oxidase
To be Published
|
|
4V3C
 
 | | The structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
6XR8
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6XT1
 
 | |
6XUZ
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV3
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 3 | | Descriptor: | 3-methyl-6-[6-[(3~{S})-3-methylmorpholin-4-yl]-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XSC
 
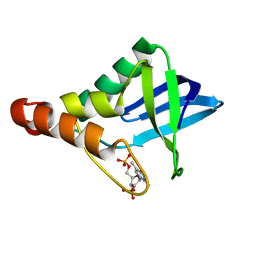 | |
4WAN
 
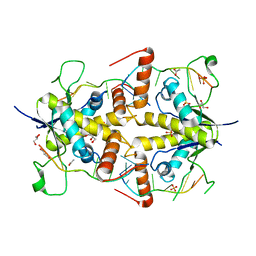 | | Crystal structure of Msl5 protein in complex with RNA at 1.8 A | | Descriptor: | ACETATE ION, Branchpoint-bridging protein, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4WBA
 
 | | Q/E mutant SA11 NSP4_CCD | | Descriptor: | GLYCEROL, Non-structural glycoprotein NSP4, PHOSPHATE ION | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
6XSG
 
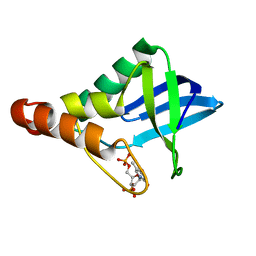 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66T at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B, Sorenson, J.L. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66T at cryogenic temperature
To be Published
|
|
6XUU
 
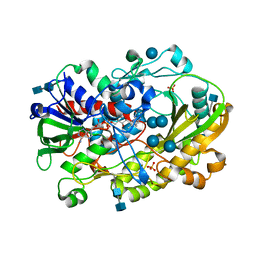 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, glucose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
4WAL
 
 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
6XYP
 
 | | Multiple system atrophy Type II-1 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
