3WO5
 
 | | Crystal structure of S147Q of Rv2613c from Mycobacterium tuberculosis | | Descriptor: | AP-4-A phosphorylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mori, S, Wachino, J, Arakawa, Y, Shibayama, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Role of Ser-147 and Ala-149 in catalytic activity of diadenosine tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv
To be Published
|
|
2ZM7
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/G181D Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
1PJC
 
 | | L-ALANINE DEHYDROGENASE COMPLEXED WITH NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (L-ALANINE DEHYDROGENASE) | | Authors: | Baker, P.J, Sawa, Y, Shibata, H, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase.
Nat.Struct.Biol., 5, 1998
|
|
1PJB
 
 | | L-ALANINE DEHYDROGENASE | | Descriptor: | L-ALANINE DEHYDROGENASE | | Authors: | Baker, P.J, Sawa, Y, Shibata, H, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of the structure and substrate binding of Phormidium lapideum alanine dehydrogenase.
Nat.Struct.Biol., 5, 1998
|
|
5WSF
 
 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5WSD
 
 | | Crystal structure of a cupin protein (tm1459) in apo form | | Descriptor: | Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5WSE
 
 | | Crystal structure of a cupin protein (tm1459) in osmium (Os) substituted form I | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
7DBH
 
 | | The mouse nucleosome structure containing H3mm18 | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2020-10-20 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
5X5G
 
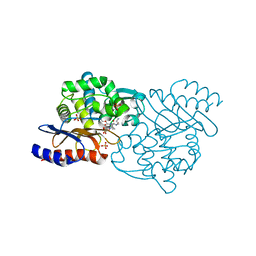 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
2IBX
 
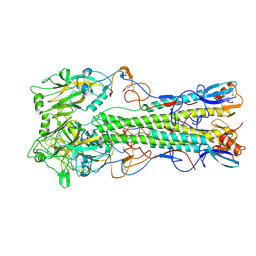 | | Influenza virus (VN1194) H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yamada, S, Russell, R.J, Gamblin, S.J, Skehel, J.J, Kawaoka, Y. | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors.
Nature, 444, 2006
|
|
5Z0A
 
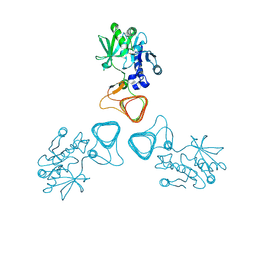 | | ST0452(Y97N)-GlcNAc binding form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual sugar-1-phosphate nucleotidylyltransferase | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
3VGV
 
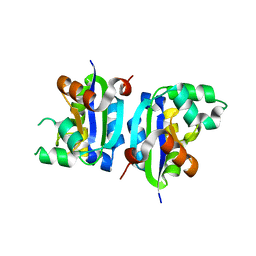 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3W2S
 
 | | EGFR kinase domain with compound4 | | Descriptor: | 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
5AY8
 
 | | Crystal structure of human nucleosome containing H3.Y | | Descriptor: | CHLORIDE ION, DNA (146-MER), H3.Y, ... | | Authors: | Kujirai, T, Horikoshi, N, Sato, K, Maehara, K, Machida, S, Osakabe, A, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of human histone H3.Y nucleosome
Nucleic Acids Res., 44, 2016
|
|
5Z09
 
 | | ST0452(Y97N)-UTP binding form | | Descriptor: | Dual sugar-1-phosphate nucleotidylyltransferase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
3W32
 
 | | EGFR kinase domain complexed with compound 20a | | Descriptor: | 4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-N-[2-(methylsulfonyl)ethyl]-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
3W2O
 
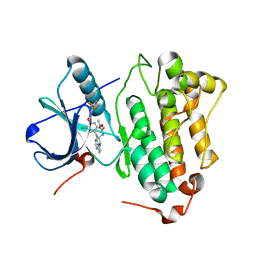 | | EGFR Kinase domain T790M/L858R Mutant with TAK-285 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-3-hydroxy-3-methylbutanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2R
 
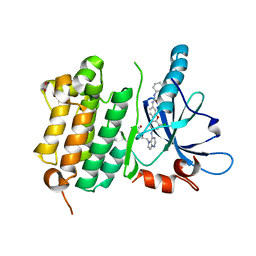 | | EGFR Kinase domain T790M/L858R mutant with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2P
 
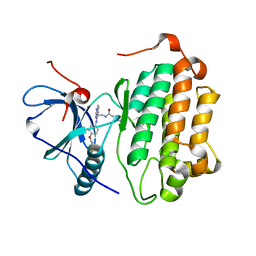 | | EGFR Kinase domain T790M/L858R mutant with compound 2 | | Descriptor: | Epidermal growth factor receptor, N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-4-(dimethylamino)butanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2Q
 
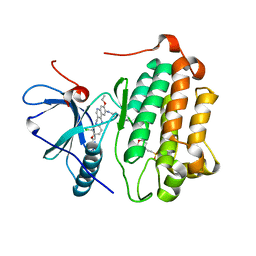 | | EGFR kinase domain T790M/L858R mutant with HKI-272 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | Authors: | Sogabe, S, Kawakita, Y, Igaki, S. | | Deposit date: | 2012-12-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W33
 
 | | EGFR kinase domain complexed with compound 19b | | Descriptor: | 4-{[4-(1-benzothiophen-4-yloxy)-3-chlorophenyl]amino}-N-(2-hydroxyethyl)-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
4BGW
 
 | | Crystal Structure of H5 (VN1194) Influenza Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, X, Coombs, P, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus.
Nature, 497, 2013
|
|
4BH3
 
 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Human Receptor Analogue 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH4
 
 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH2
 
 | | Crystal Structure of the Haemagglutinin from a Transmissible Mutant H5 Influenza Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
