7VJA
 
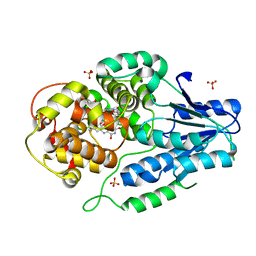 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (10 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VIX
 
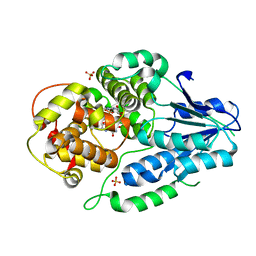 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (10 ns time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJJ
 
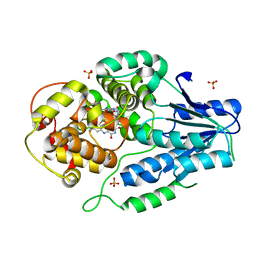 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
6LKR
 
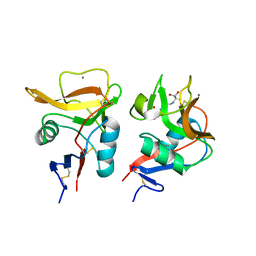 | | Crystal structure of mouse DCAR2 CRD domain complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
3W9K
 
 | | Crystal structure of thermoacidophile-specific protein STK_08120 complexed with myristic acid | | Descriptor: | FATTY ACID-BINDING PROTEIN, MYRISTIC ACID | | Authors: | Miyakawa, T, Sawano, Y, Miyazono, K, Miyauchi, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode.
J.Bacteriol., 195, 2013
|
|
4ERA
 
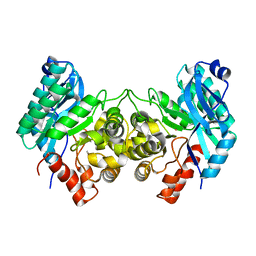 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
7WBV
 
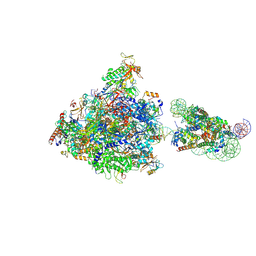 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | Descriptor: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBX
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
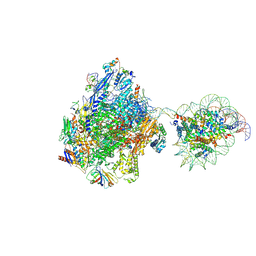 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
5GUA
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138Y mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU9
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138I mutant | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7D69
 
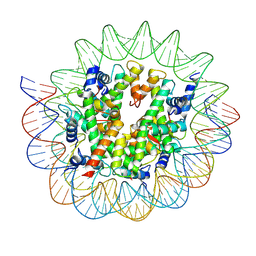 | | Cryo-EM structure of the nucleosome containing Giardia histones | | Descriptor: | 601L DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Sato, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones.
Nucleic Acids Res., 49, 2021
|
|
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | Authors: | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2Z1Q
 
 | |
6KX8
 
 | | Crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
6KX5
 
 | |
6KX4
 
 | |
3AB5
 
 | |
6I5C
 
 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
6I59
 
 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
5CPI
 
 | | Nucleosome containing unmethylated Sat2R DNA | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
5CPK
 
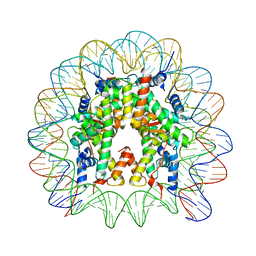 | | Nucleosome containing methylated Sat2L DNA | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
7D20
 
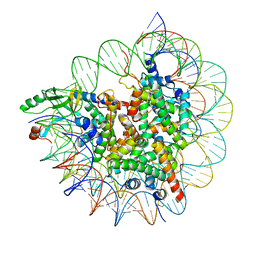 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D1Z
 
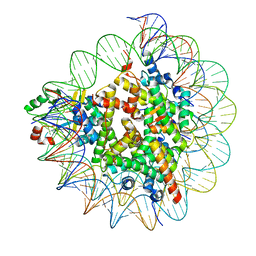 | | Cryo-EM structure of SET8-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
5CPJ
 
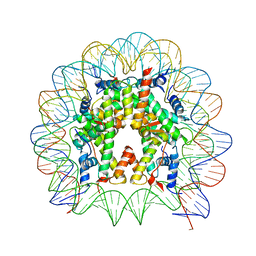 | | Nucleosome containing methylated Sat2R DNA | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
