6W6D
 
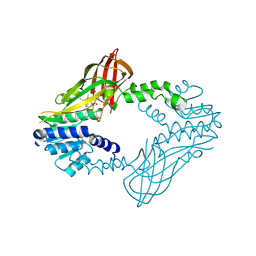 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with SGC6870 inhibitor | | Descriptor: | (5R)-4-(5-bromothiophene-2-carbonyl)-5-(3,5-dimethylphenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Jin, J, Shen, Y, Kaniskan, H.U, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A First-in-Class, Highly Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 6.
J.Med.Chem., 64, 2021
|
|
1K0H
 
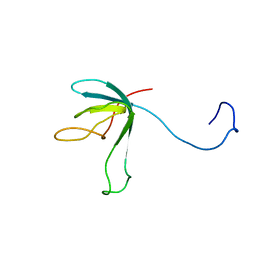 | | Solution structure of bacteriophage lambda gpFII | | Descriptor: | gpFII | | Authors: | Maxwell, K.L, Yee, A.A, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-09-19 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacteriophage lambda head-tail joining protein, gpFII.
J.Mol.Biol., 318, 2002
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
6WAJ
 
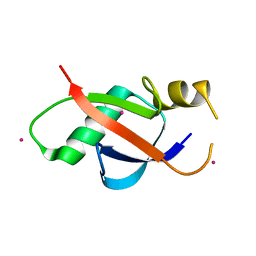 | | Crystal structure of the UBL domain of human NLE1 | | Descriptor: | NLE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the UBL domain of human NLE1
To be Published
|
|
1NOG
 
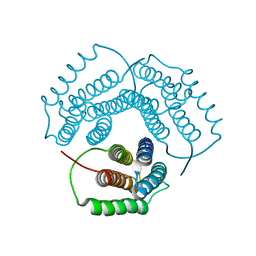 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
6WAD
 
 | | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with MT2739 inhibitor | | Descriptor: | 5-bromo-N-(diphenylmethyl)-N-methylthiophene-2-carboxamide, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Halabelian, L, Zeng, H, Dong, A, Schapira, M, De Freitas, R.F, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Protein arginine N-methyltransferase 6 (PRMT6) in complex with MT2739 inhibitor
to be published
|
|
1PES
 
 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
6VIP
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008 | | Descriptor: | TP53-binding protein 1, UNKNOWN ATOM OR ION, {4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]phenyl}(4-ethylpiperazin-1-yl)methanone | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008
to be published
|
|
1K77
 
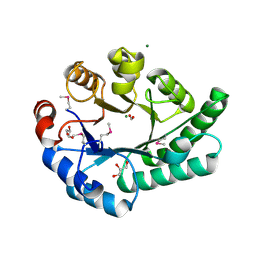 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1LV3
 
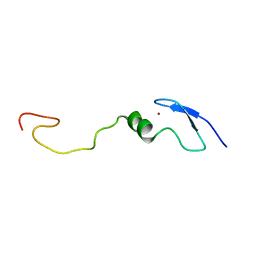 | | Solution NMR Structure of Zinc Finger Protein yacG from Escherichia coli. Northeast Structural Genomics Consortium Target ET92. | | Descriptor: | HYPOTHETICAL PROTEIN YacG, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-24 | | Release date: | 2002-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins.
Proteins, 49, 2002
|
|
6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | Descriptor: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | Authors: | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-23 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
1MP1
 
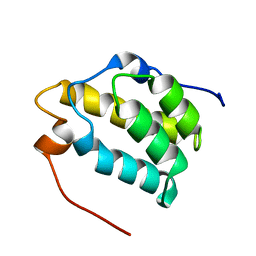 | | Solution structure of the PWI motif from SRm160 | | Descriptor: | Ser/Arg-related nuclear matrix protein | | Authors: | Szymczyna, B.R, Bowman, J, McCracken, S, Pineda-Lucena, A, Lu, Y, Cox, B, Lambermon, M, Graveley, B.R, Arrowsmith, C.H, Blencowe, B.J. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing.
Genes Dev., 17, 2003
|
|
1PU1
 
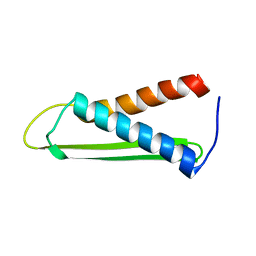 | | Solution structure of the Hypothetical protein mth677 from Methanothermobacter Thermautotrophicus | | Descriptor: | Hypothetical protein MTH677 | | Authors: | Blanco, F.J, Yee, A, Campos-Olivas, R, Devos, D, Valencia, A, Arrowsmith, C.H, Rico, M. | | Deposit date: | 2003-06-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical protein Mth677 from Methanobacterium thermoautotrophicum: A novel {alpha}+{beta} fold
Protein Sci., 13, 2004
|
|
1MV0
 
 | |
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1NYN
 
 | | Solution NMR Structure of Protein YHR087W from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YTYST425. | | Descriptor: | Hypothetical 12.0 kDa protein in NAM8-GAR1 intergenic region | | Authors: | Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-13 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1K3R
 
 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
1NE3
 
 | | Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744 | | Descriptor: | 30S ribosomal protein S28E | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J.R, Ramelot, T.A, Kennedy, M, Edwards, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.
Protein Sci., 12, 2003
|
|
1KUU
 
 | | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD | | Descriptor: | conserved protein | | Authors: | Saridakis, V, Christendat, D, Thygesen, A, Arrowsmith, C.H, Edwards, A.M, Pai, E.F. | | Deposit date: | 2002-01-22 | | Release date: | 2002-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD
PROTEINS: STRUCT.,FUNCT.,GENET., 48, 2002
|
|
1NEI
 
 | | Solution NMR Structure of Protein yoaG from Escherichia coli. Ontario Centre for Structural Proteomics Target EC0264_1_60; Northeast Structural Genomics Consortium Target ET94. | | Descriptor: | hypothetical protein yoaG | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein dimer encoded by the Yoag gene from Escherichia coli
To be published
|
|
1PET
 
 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
1LKN
 
 | |
1LFC
 
 | | BOVINE LACTOFERRICIN (LFCINB), NMR, 20 STRUCTURES | | Descriptor: | LACTOFERRICIN | | Authors: | Hwang, P.M, Zhou, N, Shan, X, Arrowsmith, C.H, Vogel, H.J. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin.
Biochemistry, 37, 1998
|
|
1LXJ
 
 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
