1X9B
 
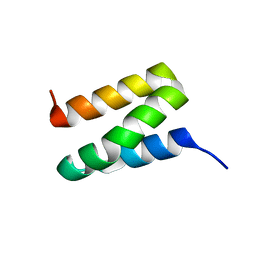 | | Solution NMR Structure of Protein Ta0354 from Thermoplasma acidophilum. Ontario Center for Structural Proteomics target TA0354_69_121; Northeast Structural Genomics Consortium Target TaT38. | | Descriptor: | hypothetical membrane protein ta0354_69_121 | | Authors: | Wu, B, Yee, A, Huang, Y.J, Ramelot, T.A, Semesi, A, Lemak, A, Edward, A, Kennedy, M, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical membrane protein ta0354_69_121 from Thermoplasma acidophilum
To be Published
|
|
1X9A
 
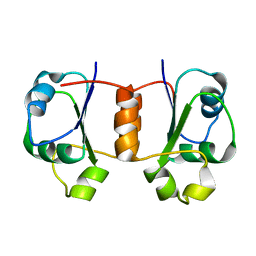 | | Solution NMR Structure of Protein Tm0979 from Thermotoga maritima. Ontario Center for Structural Proteomics Target TM0979_1_87; Northeast Structural Genomics Consortium Target VT98. | | Descriptor: | hypothetical protein TM0979 | | Authors: | Gaspar, J.A, Liu, C, Vassall, K.A, Stathopulos, P.B, Meglei, G, Stephen, R, Pineda-Lucena, A, Wu, B, Yee, A, Arrowsmith, C.H, Meiering, E.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
Protein Sci., 14, 2005
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
1NOG
 
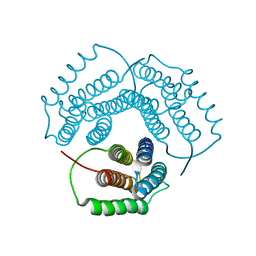 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
1LXJ
 
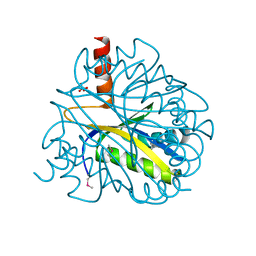 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
1MV3
 
 | |
1MUZ
 
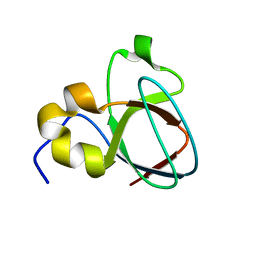 | |
1N6Z
 
 | |
1LV3
 
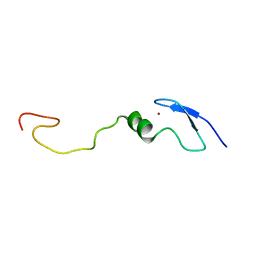 | | Solution NMR Structure of Zinc Finger Protein yacG from Escherichia coli. Northeast Structural Genomics Consortium Target ET92. | | Descriptor: | HYPOTHETICAL PROTEIN YacG, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-24 | | Release date: | 2002-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins.
Proteins, 49, 2002
|
|
1P9Q
 
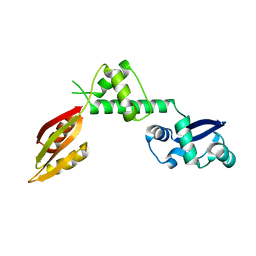 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1M94
 
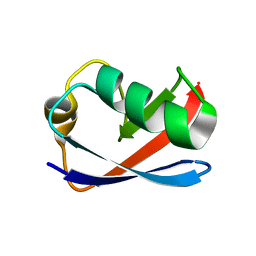 | | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1 | | Descriptor: | Protein YNR032c-a | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Edwards, A.M, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-07-26 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Yeast Ubiquitin-Like Modifier Protein Hub1
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1MP1
 
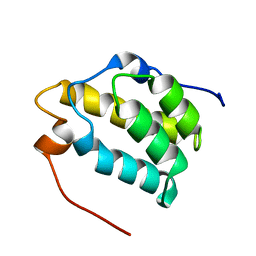 | | Solution structure of the PWI motif from SRm160 | | Descriptor: | Ser/Arg-related nuclear matrix protein | | Authors: | Szymczyna, B.R, Bowman, J, McCracken, S, Pineda-Lucena, A, Lu, Y, Cox, B, Lambermon, M, Graveley, B.R, Arrowsmith, C.H, Blencowe, B.J. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing.
Genes Dev., 17, 2003
|
|
1MV0
 
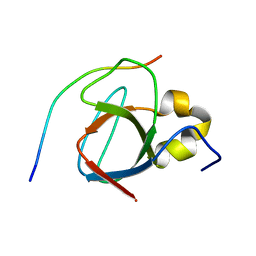 | |
1YWY
 
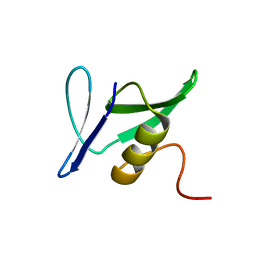 | | Solution Structure of Pseudomonas aeruginosa Protein PA2021. The Northeast Structural Genomics Consortium Target Pat85. | | Descriptor: | hypothetical protein PA2021 | | Authors: | Lin, Y.C, Liu, G, Shen, Y, Yee, A, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Pseudomonas aeruginosa Protein PA2021.
To be Published
|
|
1NEI
 
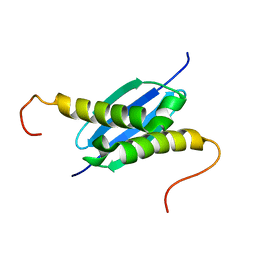 | | Solution NMR Structure of Protein yoaG from Escherichia coli. Ontario Centre for Structural Proteomics Target EC0264_1_60; Northeast Structural Genomics Consortium Target ET94. | | Descriptor: | hypothetical protein yoaG | | Authors: | Wu, B, Pineda-Lucena, A, Yee, A, Cort, J, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein dimer encoded by the Yoag gene from Escherichia coli
To be published
|
|
1LFC
 
 | | BOVINE LACTOFERRICIN (LFCINB), NMR, 20 STRUCTURES | | Descriptor: | LACTOFERRICIN | | Authors: | Hwang, P.M, Zhou, N, Shan, X, Arrowsmith, C.H, Vogel, H.J. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin.
Biochemistry, 37, 1998
|
|
1ZKH
 
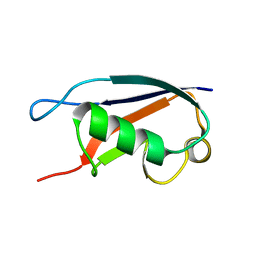 | | Solution structure of a human ubiquitin-like domain in SF3A1 | | Descriptor: | Splicing factor 3 subunit 1 | | Authors: | Lukin, J.A, Dhe-Paganon, S, Guido, V, Lemak, A, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human ubiquitin-like domain in SF3A1
To be Published
|
|
4MEQ
 
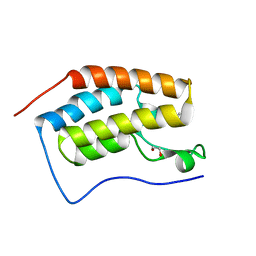 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-7-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
8SHB
 
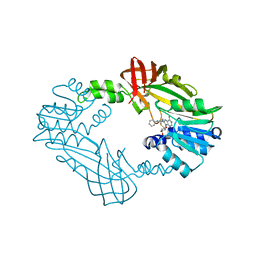 | | Crystal Structure of PRMT3 with Compound YD1-208 | | Descriptor: | 5'-S-[3-(N'-phenylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-208
To be published
|
|
4MEO
 
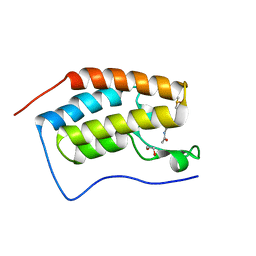 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
8SIH
 
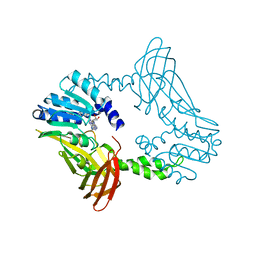 | | Crystal Structure of PRMT4 with Compound YD1-289 | | Descriptor: | 5'-{[2-(benzylcarbamamido)ethyl][3-(N'-cyclopentylcarbamimidamido)propyl]amino}-5'-deoxyadenosine, CALCIUM ION, Histone-arginine methyltransferase CARM1 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-289
To be published
|
|
8SIG
 
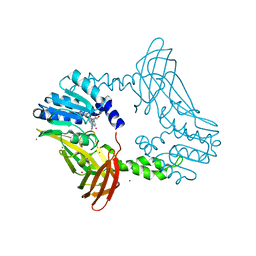 | | Crystal Structure of PRMT4 with Compound YD1-288 | | Descriptor: | 5'-{(3-aminopropyl)[2-(benzylcarbamamido)ethyl]amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, SODIUM ION, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-288
To be published
|
|
8SHR
 
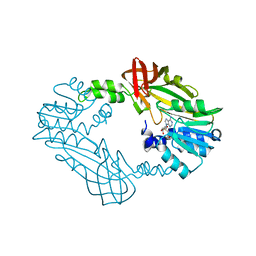 | | Crystal Structure of PRMT3 with Compound YD1-214 | | Descriptor: | 5'-S-[2-(phenylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-214
To be published
|
|
4MEP
 
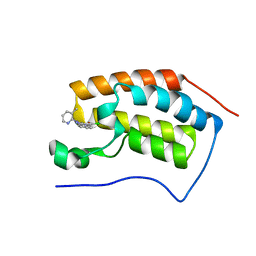 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3-chloro-pyridone ligand | | Descriptor: | 3-chloro-5-[1-(3-methylpyridin-2-yl)-3-phenyl-1H-1,2,4-triazol-5-yl]pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
