2H16
 
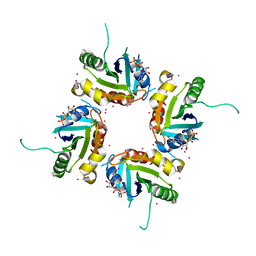 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2GRG
 
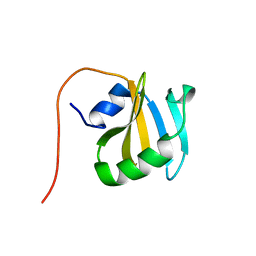 | | Solution NMR Structure of Protein YNR034W-A from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YT727; Ontario Center for Structural Proteomics Target yst6499. | | Descriptor: | hypothetical protein; Ynr034w-ap | | Authors: | Wu, B, Yee, A, Ramelot, T, Lemak, A, Semesi, A, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein yst6499 from Saccharomyces cerevisiae/ Northeast Structural Genomics Consortium Target YT727/ Ontario Center for Structural Proteomics Target yst6499
To be Published
|
|
2GSF
 
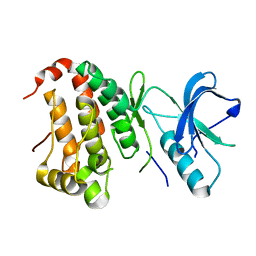 | | The Human Epha3 Receptor Tyrosine Kinase and Juxtamembrane Region | | Descriptor: | Ephrin type-A receptor 3 | | Authors: | Walker, J.R, Davis, T, Dong, A, Newman, E.M, MacKenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure Of The Human Epha3 Receptor Tyrosine Kinase and Juxtamembrane Region
To be Published
|
|
2H9L
 
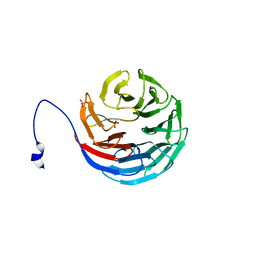 | | WDR5delta23 | | Descriptor: | SULFATE ION, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2JC6
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE 1D, N-(5-METHYL-1H-PYRAZOL-3-YL)-2-PHENYLQUINAZOLIN-4-AMINE | | Authors: | Debreczeni, J.E, Rellos, P, Fedorov, O, Niesen, F.H, Bhatia, C, Shrestha, L, Salah, E, Smee, C, Colebrook, S, Berridge, G, Gileadi, O, Bunkoczi, G, Ugochukwu, E, Pike, A.C.W, von Delft, F, Knapp, S, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase 1D
To be Published
|
|
2JIL
 
 | | Crystal structure of 2nd PDZ domain of glutamate receptor interacting protein-1 (GRIP1) | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMATE RECEPTOR INTERACTING PROTEIN-1, THIOCYANATE ION | | Authors: | Tickle, J, Elkins, J, Pike, A.C.W, Cooper, C, Salah, E, Papagrigoriou, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of 2Nd Pdz Domain of Glutamate Receptor Interacting Protein-1 (Grip1)
To be Published
|
|
2JIF
 
 | | Structure of human short-branched chain acyl-CoA dehydrogenase (ACADSB) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A PERSULFIDE, ... | | Authors: | Pike, A.C.W, Hozjan, V, Smee, C, Niesen, F.H, Kavanagh, K.L, Umeano, C, Turnbull, A.P, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Oppermann, U. | | Deposit date: | 2007-02-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Short-Branched Chain Acyl-Coa Dehydrogenase
To be Published
|
|
2JAM
 
 | | Crystal structure of human calmodulin-dependent protein kinase I G | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Debreczeni, J.E, Bullock, A, Keates, T, Niesen, F.H, Salah, E, Shrestha, L, Smee, C, Sobott, F, Pike, A.C.W, Bunkoczi, G, von Delft, F, Turnbull, A, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase I G
To be Published
|
|
2JKV
 
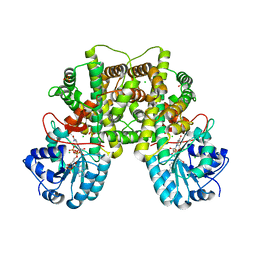 | | Structure of human Phosphogluconate Dehydrogenase in complex with NADPH at 2.53A | | Descriptor: | 6-PHOSPHOGLUCONATE DEHYDROGENASE, DECARBOXYLATING, CHLORIDE ION, ... | | Authors: | Pilka, E.S, Kavanagh, K.L, von Delft, F, Muniz, J.R.C, Murray, J, Picaud, S, Guo, K, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2008-09-01 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Structure of Human Phosphogluconate Dehydrogenase in Complex with Nadph at 2.53A
To be Published
|
|
2GUU
 
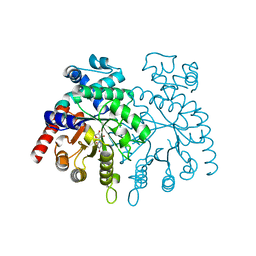 | | crystal structure of Plasmodium vivax orotidine 5-monophosphate decarboxylase with 6-aza-UMP bound | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, ODcase | | Authors: | Dong, A, Lew, J, Zhao, Y, Sundararajan, E, Wasney, G, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Pai, E.F, Kotra, L, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Plasmodium vivax orotidine 5-monophosphate decarboxylase with 6-aza-UMP bound
To be Published
|
|
2GRY
 
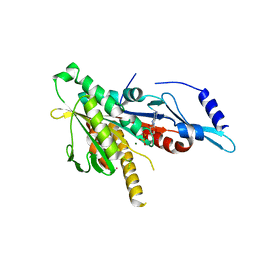 | | Crystal structure of the human KIF2 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human KIF2 motor domain in complex with ADP
to be published
|
|
2GPF
 
 | |
2GQB
 
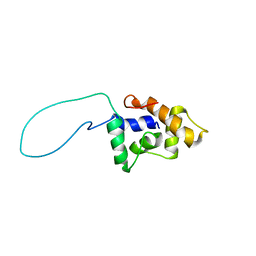 | | Solution Structure of a conserved unknown protein RPA2825 from Rhodopseudomonas palustris; (Northeast Structural Genomics Consortium Target RpT4; Ontario Centre for Structural Proteomics Target rp2812 ) | | Descriptor: | conserved hypothetical protein | | Authors: | Srisailam, S, Lukin, J.A, Yee, A, Lemak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a conserved unknown protein RPA2825 from Rhodopseudomonas palustris
To be Published
|
|
2JIN
 
 | | Crystal structure of PDZ domain of Synaptojanin-2 binding protein | | Descriptor: | SODIUM ION, SULFATE ION, SYNAPTOJANIN-2 BINDING PROTEIN | | Authors: | Tickle, J, Phillips, C, Pike, A.C.W, Cooper, C, Salah, E, Elkins, J, Turnbull, A.P, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Pdz Domain of Synaptojanin-2 Binding Protein
To be Published
|
|
2JNE
 
 | | NMR structure of E.coli YfgJ modelled with two Zn+2 bound. Northeast Structural Genomics Consortium Target ER317. | | Descriptor: | Hypothetical protein yfgJ, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Semesi, A, Lukin, J.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of E.coli YfgJ bound to two Zn+2.
To be Published
|
|
2JPF
 
 | |
2JRA
 
 | | A novel domain-swapped solution NMR structure of protein RPA2121 from Rhodopseudomonas palustris. Northeast Structural Genomics Target RpT6 | | Descriptor: | Protein RPA2121 | | Authors: | Wu, B, Yee, A, Lemak, A, Cort, J, Bansal, S, Semest, A, Guido, V, Kennedy, M.A, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-21 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A novel domain-swapped solution NMR structure of protein RPA2121 from Rhodopseudomonas palustris.
To be Published
|
|
2JO6
 
 | | NMR structure of the E.coli protein NirD, Northeast Structural Genomics target ET100 | | Descriptor: | Nitrite reductase [NAD(P)H] small subunit | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Guido, V, Lukin, J.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of E.coli NirD.
To be Published
|
|
2JPI
 
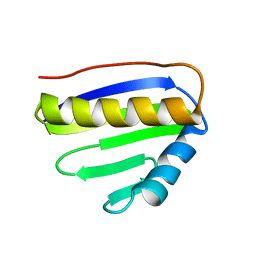 | | NMR structure of PA4090 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical protein | | Authors: | Ai, X, Semesi, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-05-13 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | chemical shift assignments of PA4090 from Pseudomonas aeruginosa
To be Published
|
|
2KI8
 
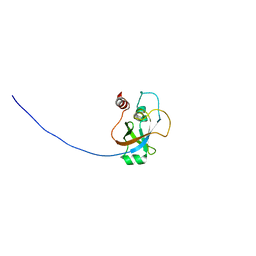 | | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium target AtT7 | | Descriptor: | Tungsten formylmethanofuran dehydrogenase, subunit D (FwdD-2) | | Authors: | Eletsky, A, Wu, Y, Yee, A, Fares, C, Lee, H.W, Arrowsmith, C.H, Prestegard, J.H, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus
To be Published
|
|
6V2S
 
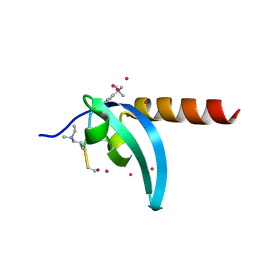 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2H
 
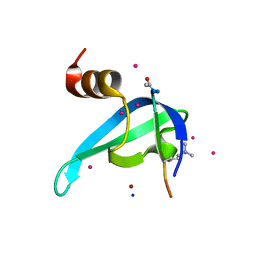 | | Crystal structure of CDYL2 in complex with H3tK27me3 | | Descriptor: | Chromodomain Y-like protein 2, H3tK27me3, NICKEL (II) ION, ... | | Authors: | Dong, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6VDB
 
 | | SETD2 in complex with a H3-variant super-substrate peptide | | Descriptor: | ALA-PRO-ARG-PHE-GLY-GLY-VAL-MET-ARG-PRO-ASN-ARG, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Beldar, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Jeltsch, A, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence specificity analysis of the SETD2 protein lysine methyltransferase and discovery of a SETD2 super-substrate.
Commun Biol, 3, 2020
|
|
8RU1
 
 | | Chromatin remodeling regulator CECR2 with in crystallo disulfide bond | | Descriptor: | Chromatin remodeling regulator CECR2, GLYCEROL, SODIUM ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
6V3N
 
 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
