5G4P
 
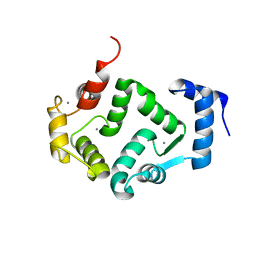 | | Crystal structure of human hippocalcin at 2.4 A resolution | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Antonyuk, S.V, Helassa, N, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-05-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
5G58
 
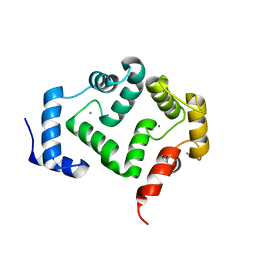 | | Crystal structure of A190T mutant of human hippocalcin AT 2.5 A resolution | | Descriptor: | CALCIUM ION, Neuron-specific calcium-binding protein hippocalcin | | Authors: | Antonyuk, S.V, Helassa, N, Lian, L.Y, Haynes, L.P, Burgoyne, R.D. | | Deposit date: | 2016-05-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Biophysical and functional characterization of hippocalcin mutants responsible for human dystonia.
Hum. Mol. Genet., 26, 2017
|
|
6EU8
 
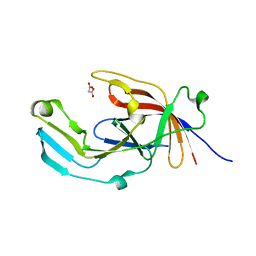 | |
8B61
 
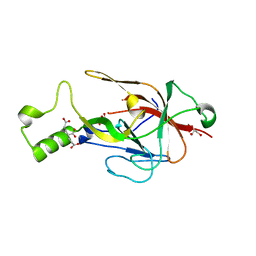 | | Crystal structure of BfrC protein from Bacteroides fragilis NCTC 9343 | | Descriptor: | Conserved hypothetical lipoprotein, GLYCEROL, pentane-1,3,5-tricarboxylic acid | | Authors: | Antonyuk, S.V, Barnett, K, Strange, R.W, Olczak, T. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes.
Faseb J., 37, 2023
|
|
8B6A
 
 | |
6EWM
 
 | | Crystal structure of heme free PORPHYROMONAS GINGIVALIS HEME-BINDING PROTEIN HMUY | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Haemophore HmuY, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Bielecki, M, Olczak, T, Olczak, M. | | Deposit date: | 2017-11-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tannerella forsythiaTfo belongs toPorphyromonas gingivalisHmuY-like family of proteins but differs in heme-binding properties.
Biosci. Rep., 38, 2018
|
|
6FJA
 
 | |
6F1Q
 
 | |
6R2H
 
 | | Crystal structure of Apo PinO from Porphyromonas gingivitis | | Descriptor: | GLYCEROL, HmuY protein | | Authors: | Antonyuk, S.V, Bielecki, M, Strange, R.W, Capper, M, Olczak, T, Olczak, M. | | Deposit date: | 2019-03-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode.
Biochem.J., 477, 2020
|
|
2XX1
 
 | | STRUCTURE OF THE N90S MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2XX0
 
 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2XWZ
 
 | | STRUCTURE OF THE RECOMBINANT NATIVE NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | ACETATE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
8CCX
 
 | | Human SOD1 in complex with S-XL6 cross-linker | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-01-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
2JLP
 
 | | Crystal structure of human extracellular copper-zinc superoxide dismutase. | | Descriptor: | COPPER (II) ION, EXTRACELLULAR SUPEROXIDE DISMUTASE (CU-ZN), THIOCYANATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Marklund, S.L, Hasnain, S.S. | | Deposit date: | 2008-09-14 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Human Extracellular Copper-Zinc Superoxide Dismutase at 1.7 A Resolution: Insights Into Heparin and Collagen Binding.
J.Mol.Biol., 388, 2009
|
|
3IXQ
 
 | | Structure of ribose 5-phosphate isomerase a from methanocaldococcus jannaschii | | Descriptor: | ACETATE ION, CHLORIDE ION, Ribose-5-phosphate isomerase A, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of an archaeal ribose-5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3IWT
 
 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KB6
 
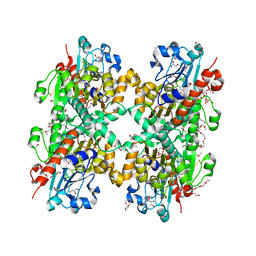 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6XY3
 
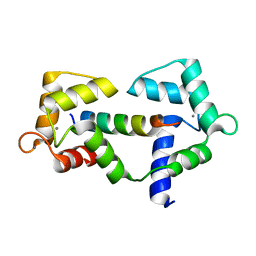 | |
6XXF
 
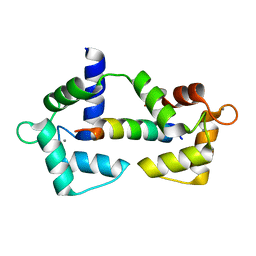 | |
6XXX
 
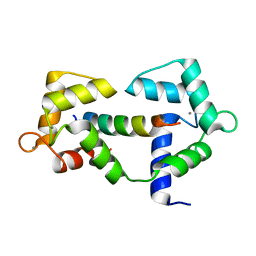 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
1OBG
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHORIBOSYLAMIDOIMIDAZOLE- SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBD
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBF
 
 | | The crystal structure of Glyceraldehyde 3-phosphate Dehydrogenase from Alcaligenes xylosoxidans at 1.7A resolution. | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Eady, R.R, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Glyceraldehyde 3-Phosphate Dehydrogenase from Alcaligenes Xylosoxidans at 1.7 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6QQ2
 
 | | Crystal structure of nitrite bound Y323F mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QQ1
 
 | | Crystal structure of as isolated Y323F mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
