5YTH
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3P2T
 
 | | Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, SULFATE ION | | Authors: | Chen, Y, Nam, G, Cheng, H, Zhang, J.H, Willcox, B.E, Gao, G.F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance
J.Biol.Chem., 286, 2011
|
|
5Y22
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV | | Descriptor: | 22AA-PSTD peptide | | Authors: | Lu, B, Liao, S.M, Huang, J.M, Lu, Z.L, Chen, D, Liu, X.H, Zhou, G.P, Huang, R.B. | | Deposit date: | 2017-07-23 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV
To Be Published
|
|
4AF3
 
 | | Human Aurora B Kinase in complex with INCENP and VX-680 | | Descriptor: | AURORA KINASE B, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, INNER CENTROMERE PROTEIN | | Authors: | Elkins, J.M, Vollmar, M, Wang, J, Picaud, S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
J.Med.Chem., 55, 2012
|
|
1NPE
 
 | | Crystal structure of Nidogen/Laminin Complex | | Descriptor: | CADMIUM ION, Laminin gamma-1 chain, nidogen | | Authors: | Takagi, J, Yang, Y.T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 2003-01-17 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex between nidogen and laminin fragments reveals a paradigmatic
beta-propeller interface
Nature, 424, 2003
|
|
2WV5
 
 | | Crystal structure of foot-and-mouth disease virus 3C protease in complex with a decameric peptide corresponding to the VP1-2A cleavage junction with a GLN to Glu substitution at P1 | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS (SEROTYPE A) VARIANT VP1 CAPSID PROTEIN, PICORNAIN 3C | | Authors: | Zunszain, P.A, Knox, S.R, Sweeney, T.R, Yang, J, Roque-Rosell, N, Belsham, G.J, Leatherbarrow, R.J, Curry, S. | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
J.Mol.Biol., 395, 2010
|
|
2LVZ
 
 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
2WV4
 
 | | Crystal structure of foot-and-mouth disease virus 3C protease in complex with a decameric peptide corresponding to the VP1-2A cleavage junction | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS (SEROTYPE A) VARIANT VP1 CAPSID PROTEIN, PICORNAIN 3C | | Authors: | Zunszain, P.A, Knox, S.R, Sweeney, T.R, Yang, J, Roque-Rosell, N, Belsham, G.J, Leatherbarrow, R.J, Curry, S. | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
J.Mol.Biol., 395, 2010
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
2Y1R
 
 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
1WBZ
 
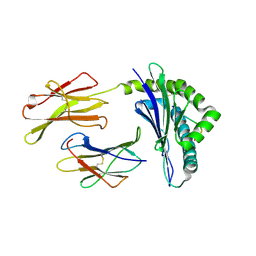 | | CRYSTAL STRUCTURES OF MURINE MHC CLASS I H-2 Db AND Kb MOLECULES IN COMPLEX WITH CTL EPITOPES FROM INFLUENZA A VIRUS: IMPLICATIONS FOR TCR REPERTOIRE SELECTION AND IMMUNODOMINANCE | | Descriptor: | BETA-2MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, K-B ALPHA CHAIN PRECURSOR, ... | | Authors: | Meijers, R, Lai, C, Yang, Y, Liu, J, Zhong, W, Wang, J, Reinherz, E.L. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Murine Mhc Class I H-2 D(B) and K(B) Molecules in Complex with Ctl Epitopes from Influenza a Virus: Implications for Tcr Repertoire Selection and Immunodominance
J.Mol.Biol., 345, 2005
|
|
3SCX
 
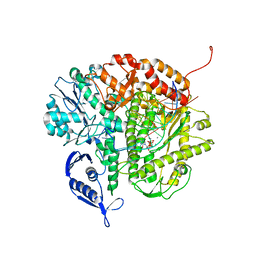 | | RB69 DNA Polymerase Triple Mutant(L561A/S565G/Y567A) Ternary Complex with dUpNpp and a Deoxy-terminated Primer in the Presence of Ca2+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-06-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
1WBX
 
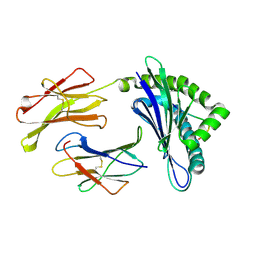 | | CRYSTAL STRUCTURES OF MURINE MHC CLASS I H-2 Db AND Kb MOLECULES IN COMPLEX WITH CTL EPITOPES FROM INFLUENZA A VIRUS: IMPLICATIONS FOR TCR REPERTOIRE SELECTION AND IMMUNODOMINANCE | | Descriptor: | BETA-2MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Meijers, R, Lai, C, Yang, Y, Liu, J, Zhong, W, Wang, J, Reinherz, E.L. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Murine Mhc Class I H-2 D(B) and K(B) Molecules in Complex with Ctl Epitopes from Influenza a Virus: Implications for Tcr Repertoire Selection and Immunodominance
J.Mol.Biol., 345, 2005
|
|
7WKC
 
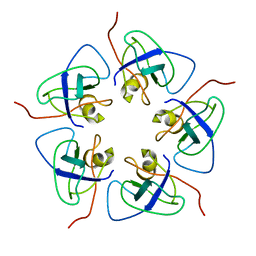 | |
2XCO
 
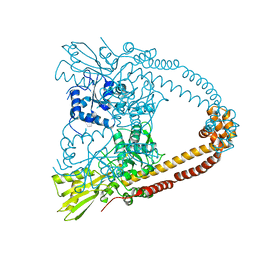 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
7X68
 
 | | CYS179 and CYS504 of CRMP2 were covalently binded by a Sesquiterpene lactone | | Descriptor: | (3aR,5S,8R,8aR,9aR)-5,8a-dimethyl-3-methylidene-8-oxidanyl-5,6,7,8,9,9a-hexahydro-3aH-benzo[f][1]benzofuran-2-one, Dihydropyrimidinase-related protein 2, SODIUM ION | | Authors: | Zhang, S.D, Ma, Y.F, Zhang, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CYS179 and CYS504 of CRMP2 were covalently binded by a Sesquiterpene lactone
To Be Published
|
|
2XKJ
 
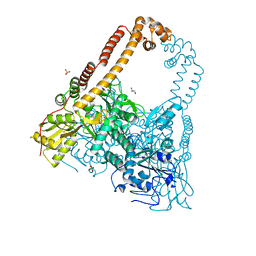 | | CRYSTAL STRUCTURE OF CATALYTIC CORE OF A. BAUMANNII TOPO IV (PARE- PARC FUSION TRUNCATE) | | Descriptor: | GLYCEROL, SULFATE ION, TOPOISOMERASE IV | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
4N77
 
 | |
2M2R
 
 | | Solution structure of MCh-2: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-2 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2XKK
 
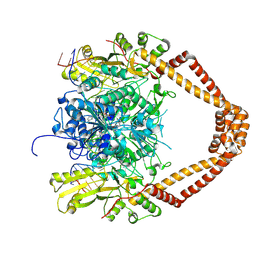 | | CRYSTAL STRUCTURE OF MOXIFLOXACIN, DNA, and A. BAUMANNII TOPO IV (PARE-PARC FUSION TRUNCATE) | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA, MAGNESIUM ION, ... | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
4MP2
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PA1 | | Descriptor: | (5-bromo-2,4-dihydroxyphenyl)(1,3-dihydro-2H-isoindol-2-yl)methanone, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4NN2
 
 | | Protein Crystal Structure of Human Borjeson-Forssman-Lehmann Syndrome Associated Protein PHF6 | | Descriptor: | GLYCEROL, PHD finger protein 6, ZINC ION | | Authors: | Liu, Z, Li, F, Zhang, J, Mei, Y, Wu, J, Shi, Y. | | Deposit date: | 2013-11-16 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Crystal Structure of the second extended PHD domain of human PHF6 protein
J.Biol.Chem., 2014
|
|
5ZWV
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29 | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W. | | Deposit date: | 2018-05-17 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5EA6
 
 | | Crystal Structure of Inhibitor BTA-9881 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | (9~{b}~{R})-9~{b}-(4-chlorophenyl)-1-pyridin-3-ylcarbonyl-2,3-dihydroimidazo[5,6]pyrrolo[1,2-~{a}]pyridin-5-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
