1WAF
 
 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
1WAJ
 
 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
2FAP
 
 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-(C16)-ETHOXY RAPAMYCIN COMPLEX INTERACTING WITH HUMA | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FRAP | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2XMS
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | CHLORIDE ION, IMIDAZOLE, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
2XMR
 
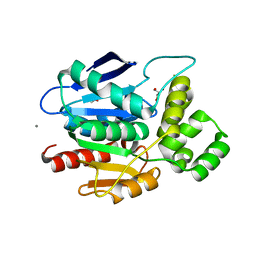 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
1HQY
 
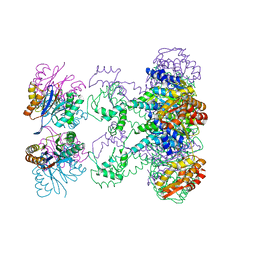 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1HT1
 
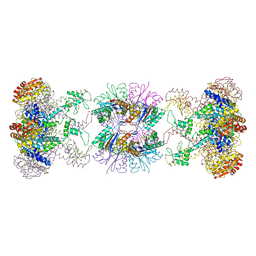 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
2K0C
 
 | | Zinc-finger 2 of Nup153 | | Descriptor: | Nuclear pore complex protein Nup153, ZINC ION | | Authors: | Bangert, J.A, Schrader, N, Vetter, I.R, Stoll, R. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex.
Structure, 16, 2008
|
|
8R1X
 
 | |
2ZIW
 
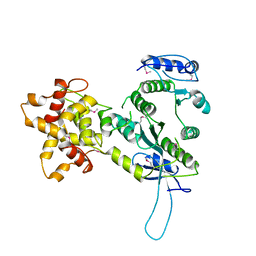 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
6H1X
 
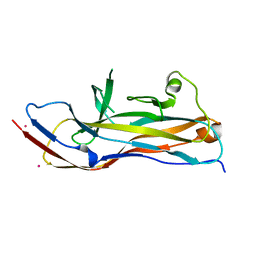 | |
1JOR
 
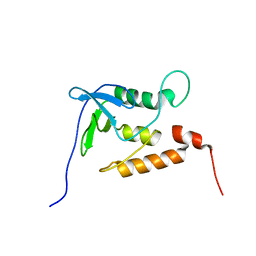 | | Ensemble structures for unligated Staphylococcal nuclease-H124L | | Descriptor: | staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
2ZIU
 
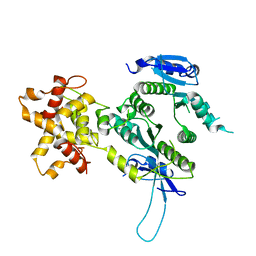 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIV
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIX
 
 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
1HT2
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
3A7C
 
 | | Crystal structure of TLR2-PE-DTPA complex | | Descriptor: | (10S,13R)-3-{2-[{2-[bis(carboxymethyl)amino]ethyl}(carboxymethyl)amino]ethyl}-10-hydroxy-5,16-dioxo-13-(tetradecanoyloxy)-9,11,15-trioxa-3,6-diaza-10-phosphanonacosan-1-oic acid 10-oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kang, J.Y, Jin, M.S, Lee, J.-O. | | Deposit date: | 2009-09-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer
Immunity, 31, 2009
|
|
1JOO
 
 | | Averaged structure for unligated Staphylococcal nuclease-H124L | | Descriptor: | staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
1JOK
 
 | | Averaged structure for Staphylococcal nuclease-H124L in ternary complex with Ca2+ and thymidine-3',5'-bisphosphate | | Descriptor: | THYMIDINE-3',5'-DIPHOSPHATE, staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
1JOQ
 
 | | Ensemble structures for Staphylococcal nuclease-H124L in ternary complex with Ca2+ and thymidine-3',5'-bisphosphate | | Descriptor: | THYMIDINE-3',5'-DIPHOSPHATE, staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
1JL4
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE | | Descriptor: | H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, A-K ALPHA CHAIN, A-K BETA CHAIN, ... | | Authors: | Wang, J.-H, Meijers, R, Reinherz, E.L. | | Deposit date: | 2001-07-15 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KP8
 
 | |
1ZA1
 
 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of CTP at 2.20 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, J, Stieglitz, K.A, Cardia, J.P, Kantrowitz, E.R. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
