8PYI
 
 | | Human IGF1R with inhibitor 6 | | Descriptor: | 3-[8-azanyl-1-(4-ethoxy-8-fluoranyl-2-phenyl-quinolin-7-yl)imidazo[1,5-a]pyrazin-3-yl]-1-methyl-cyclobutan-1-ol, Insulin-like growth factor 1 receptor beta chain | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Human IGF1R with inhibitor 6
To Be Published
|
|
8PYJ
 
 | | Human IGF1R with inhibitor 8 | | Descriptor: | 5,5-dimethyl-1-(quinolin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Human IGF1R with inhibitor 8
To Be Published
|
|
8PYK
 
 | | Human IGF1R with inhibitor 47 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, Insulin-like growth factor 1 receptor beta chain, NICKEL (II) ION | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human IGF1R with inhibitor 47
To Be Published
|
|
8PYL
 
 | | Human IGF1R with inhibitor 53 | | Descriptor: | (5S)-5-methyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Human IGF1R with inhibitor 53
To Be Published
|
|
8PYN
 
 | | Human IGF1R with inhibitor 56 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfonyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Human IGF1R with inhibitor 56
To Be Published
|
|
8PYM
 
 | | Human IGF1R with inhibitor 54 | | Descriptor: | 5,5-dimethyl-1-(1H-pyrrolo[2,3-b]pyridin-4-ylmethyl)-3-[4-(trifluoromethylsulfanyl)phenyl]imidazolidine-2,4-dione, CADMIUM ION, Insulin-like growth factor 1 receptor beta chain, ... | | Authors: | Dreyer, M.K, Wang, J, Elkins, J.M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Human IGF1R with inhibitor 54
To Be Published
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
5Z5K
 
 | | Structure of the DCC-Draxin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Draxin, ... | | Authors: | Liu, Y, Xiao, J, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2018-06-20 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
3CMZ
 
 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
6JPK
 
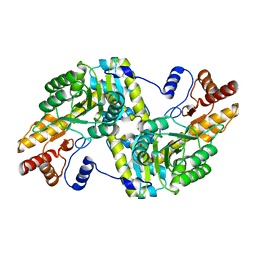 | |
3IZI
 
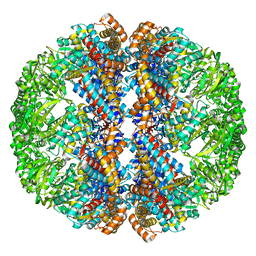 | | Mm-cpn rls with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZH
 
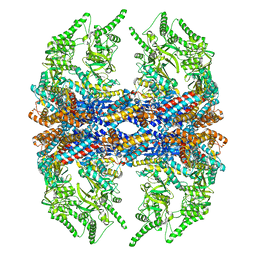 | | Mm-cpn D386A with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
6JJT
 
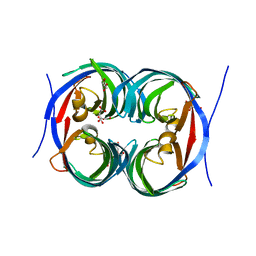 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
3IZM
 
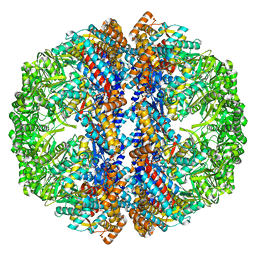 | | Mm-cpn wildtype with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZN
 
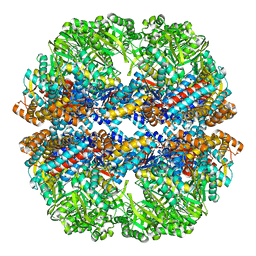 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZL
 
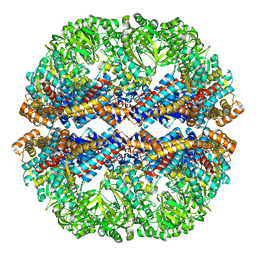 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3J1T
 
 | | High affinity dynein microtubule binding domain - tubulin complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, seryl t-RNA synthetase chimera, Tubulin alpha-1B chain, ... | | Authors: | Redwine, W.B, Hernandez-Lopez, R, Zou, S, Huang, J, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2012-06-25 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structural basis for microtubule binding and release by dynein.
Science, 337, 2012
|
|
4CDX
 
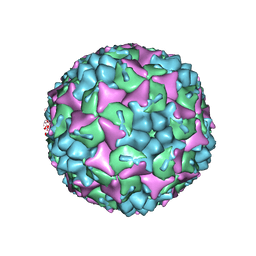 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4GSB
 
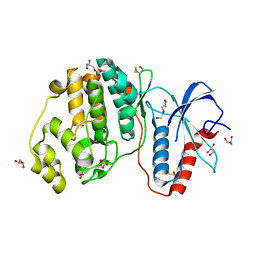 | |
4ZJS
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia Californica (Ac-AChBP) containing the main immunogenic region (MIR) from the human alpha 1 subunit of the muscle nicotinic acetylcholine receptor in complex with anatoxin-A. | | Descriptor: | 1-[(1R,6R)-9-azabicyclo[4.2.1]non-2-en-2-yl]ethanone, Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor,Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J, Park, J.F, Luo, J, Lindsatrom, J, Taylor, P. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2301 Å) | | Cite: | Main immunogenic region structure promotes binding of conformation-dependent myasthenia gravis autoantibodies, nicotinic acetylcholine receptor conformation maturation, and agonist sensitivity.
J. Neurosci., 29, 2009
|
|
4RNY
 
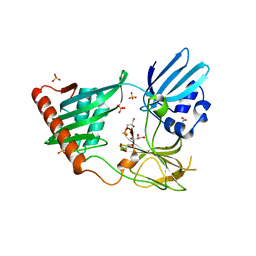 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5L4M
 
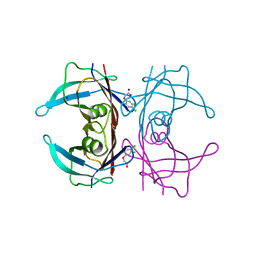 | | Crystal Structure of Human Transthyretin in Complex with 3,5,6-Trichloro-2-pyridinyloxyacetic acid (Triclopyr) | | Descriptor: | SODIUM ION, Transthyretin, Triclopyr | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
5ZM7
 
 | | Crystal structure of ORP1-ORD in complex with cholesterol at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
