8UPT
 
 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
7EK4
 
 | | prawn ferritin to coordinate with heavy metal ions | | Descriptor: | FE (III) ION, Ferritin, MERCURY (II) ION | | Authors: | Wang, Y, Zang, J. | | Deposit date: | 2021-04-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights for the Stronger Ability of Shrimp Ferritin to Coordinate with Heavy Metal Ions as Compared to Human H-Chain Ferritin.
Int J Mol Sci, 22, 2021
|
|
7EK7
 
 | | prawn ferritin to coordinate with heavy metal ions | | Descriptor: | Ferritin, MERCURY (II) ION | | Authors: | Wang, Y, Zang, J. | | Deposit date: | 2021-04-04 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights for the Stronger Ability of Shrimp Ferritin to Coordinate with Heavy Metal Ions as Compared to Human H-Chain Ferritin.
Int J Mol Sci, 22, 2021
|
|
7EK5
 
 | | prawn ferritin to coordinate with heavy metal ions | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin | | Authors: | Wang, Y, Zang, J. | | Deposit date: | 2021-04-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights for the Stronger Ability of Shrimp Ferritin to Coordinate with Heavy Metal Ions as Compared to Human H-Chain Ferritin.
Int J Mol Sci, 22, 2021
|
|
5YEF
 
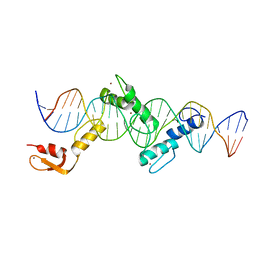 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | Descriptor: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YKW
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | Thioredoxin-3, mitochondrial, peptide THR-PRO-VAL-CYS-THR-THR-GLU-VAL | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
to be published
|
|
5NUF
 
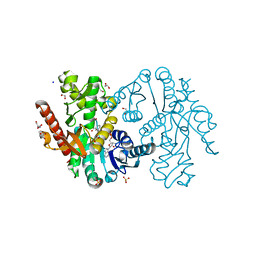 | | Cytosolic Malate Dehydrogenase 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
5YR2
 
 | | Structure of cpGFP66BPA | | Descriptor: | Green fluorescent protein | | Authors: | Wang, L, Kang, F, Wang, J. | | Deposit date: | 2017-11-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure of cpGFP66BPA
To Be Published
|
|
5NUE
 
 | | Cytosolic Malate Dehydrogenase 1 (peroxide-treated) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ETHOXYETHANOL, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.35000277 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
4F5Y
 
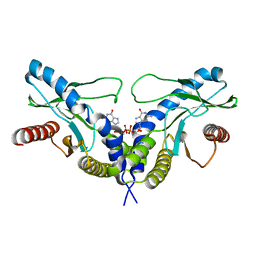 | | Crystal structure of human STING CTD complex with C-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
8J25
 
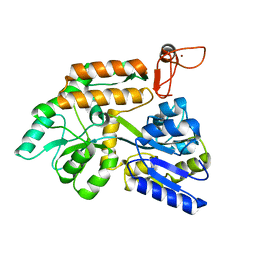 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
5Z96
 
 | | Structure of the mouse TRPC4 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-02-02 | | Release date: | 2018-04-18 | | Last modified: | 2018-08-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mouse TRPC4 ion channel.
Nat Commun, 9, 2018
|
|
6J99
 
 | |
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
2LI6
 
 | |
5JJA
 
 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
8HQI
 
 | |
8HQH
 
 | |
8HQJ
 
 | |
7QA4
 
 | |
5JRE
 
 | | Crystal structure of NeC3PO in complex with ssDNA. | | Descriptor: | 9-METHYL-9H-PURIN-6-AMINE, ADENINE, NEQ131, ... | | Authors: | Gan, J, Zhang, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
4UIF
 
 | | Cryo-EM structure of Dengue virus serotype 2 in complex with antigen-binding fragments of human antibody 2D22 | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN PVP94 07 - ENVELOPE PROTEIN, ... | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.-N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
8SXO
 
 | |
4UML
 
 | | Crystal structure of ganglioside induced differentiation associated protein 2 (GDAP2) macro domain | | Descriptor: | GANGLIOSIDE-INDUCED DIFFERENTIATION-ASSOCIATED PROTEIN 2 | | Authors: | Elkins, J.M, Wang, J, Kopec, J, Wang, D, Strain-Damerell, C, Shrestha, L, Sieg, C, Tallant, C, Newman, J.A, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2014-05-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Gdap2
To be Published
|
|
