5GR8
 
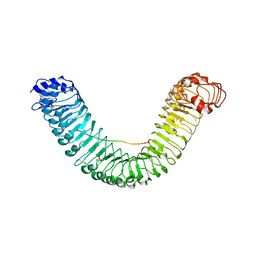 | | Crystal structure of PEPR1-AtPEP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Elicitor peptide 1, ... | | Authors: | Chai, J.J, Tang, J. | | Deposit date: | 2016-08-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1
Cell Res., 25, 2015
|
|
6JJS
 
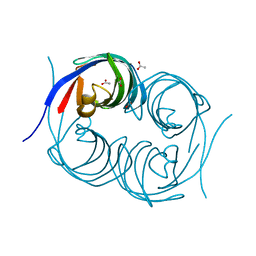 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
7WC2
 
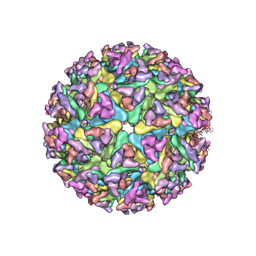 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Spike glycoprotein E1, Spike glycoprotein E2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
6GGY
 
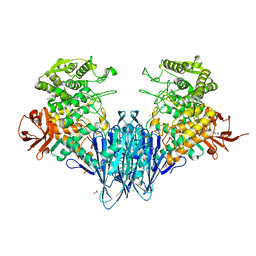 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with sulphate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Laminaribiose phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
4FK2
 
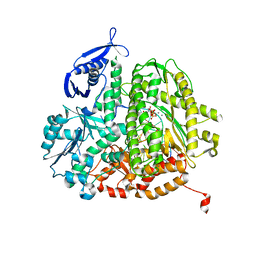 | | RB69 DNA polymerase ternary complex with dTTP/dG | | Descriptor: | CALCIUM ION, DNA polymerase, DNA primer, ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2012-06-12 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase.
J.Am.Chem.Soc., 135, 2013
|
|
5V9P
 
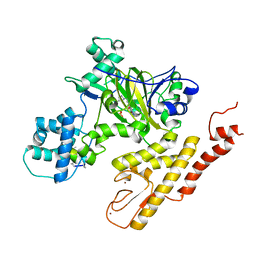 | | Crystal structure of pyrrolidine amide inhibitor [(3S)-3-(4-bromo-1H-pyrazol-1-yl)pyrrolidin-1-yl][3-(propan-2-yl)-1H-pyrazol-5-yl]methanone (compound 35) in complex with KDM5A | | Descriptor: | Lysine-specific demethylase 5A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2LBR
 
 | |
4ORA
 
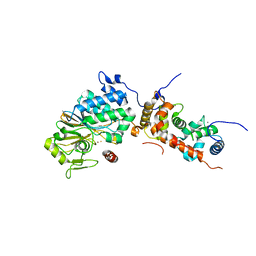 | | Crystal structure of a human calcineurin mutant | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
6GH3
 
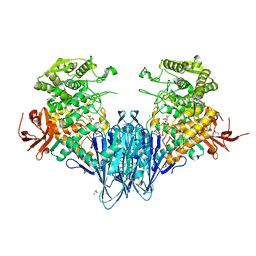 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-man-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
4OBY
 
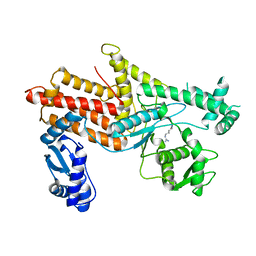 | | Crystal Structure of E.coli Arginyl-tRNA Synthetase and Ligand Binding Studies Revealed Key Residues in Arginine Recognition | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Bi, K, Zheng, Y, Dong, J, Gao, F, Wang, J, Wang, Y, Gong, W. | | Deposit date: | 2014-01-08 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Crystal structure of E. coli arginyl-tRNA synthetase and ligand binding studies revealed key residues in arginine recognition.
Protein Cell, 5, 2014
|
|
5Y0H
 
 | | Solution structure of arenicin-3 derivative N6 | | Descriptor: | N6 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
4J2D
 
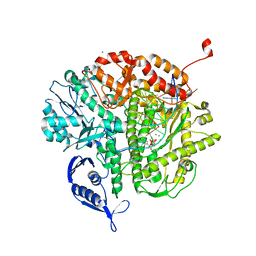 | | RB69 DNA Polymerase L415K Ternary Complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Role of L415 in the Replication of RB69 DNA Polymerase
To be Published
|
|
2LBQ
 
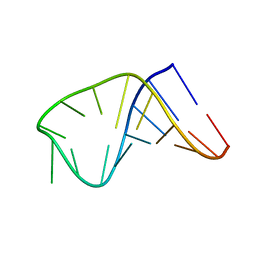 | | NMR structure of i6A37_tyrASL | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*(6IA)P*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
5Y0J
 
 | | Solution structure of arenicin-3 derivative N2 | | Descriptor: | N2 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
1T0P
 
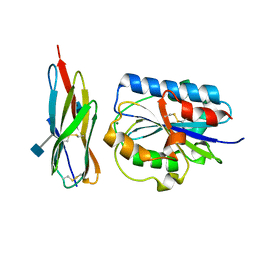 | | Structural Basis of ICAM recognition by integrin alpahLbeta2 revealed in the complex structure of binding domains of ICAM-3 and alphaLbeta2 at 1.65 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, Intercellular adhesion molecule-3, ... | | Authors: | Song, G, Yang, Y.T, Liu, J.H, Shimaoko, M, Springer, T.A, Wang, J.H. | | Deposit date: | 2004-04-12 | | Release date: | 2005-03-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alphaLbeta2.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2LUA
 
 | | Solution structure of CXC domain of MSL2 | | Descriptor: | Protein male-specific lethal-2, ZINC ION | | Authors: | Feng, Y, Ye, K, Zheng, S, Wang, J. | | Deposit date: | 2012-06-09 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MSL2 CXC Domain Reveals an Unusual Zn(3)Cys(9) Cluster and Similarity to Pre-SET Domains of Histone Lysine Methyltransferases.
Plos One, 7, 2012
|
|
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
3LM0
 
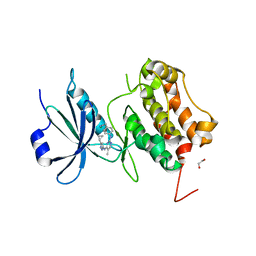 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Serine/threonine-protein kinase 17B, ... | | Authors: | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: |
|
|
5N9Q
 
 | | Structure of A. thaliana RCD1(468-567) | | Descriptor: | Inactive poly [ADP-ribose] polymerase RCD1 | | Authors: | Tossavainen, H, Hellman, M, Vainonen, J, Kangasjarvi, J, Permi, P. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors.
Elife, 8, 2019
|
|
2BN5
 
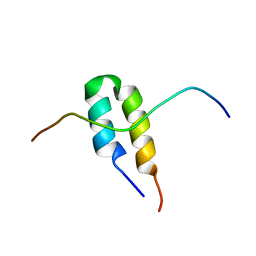 | | P-Element Somatic Inhibitor Protein Complex with U1-70k proline-rich peptide | | Descriptor: | PSI, U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA | | Authors: | Ignjatovic, T, Yang, J.-C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
4IXL
 
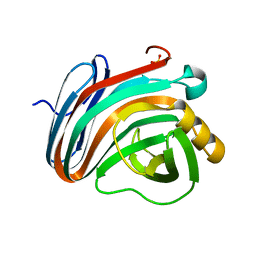 | | Crystal structure of endo-beta-1,4-xylanase from the alkaliphilic Bacillus sp. SN5 | | Descriptor: | SULFATE ION, xylanase | | Authors: | Bai, W.Q, Xue, Y.F, Huang, J.X, Zhou, C, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-01-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure Analysis of Alkaline xylanase from Alkaliphilic Bacillus sp. SN5 Implications for pH-dependent enzyme activity
To be Published
|
|
7WQI
 
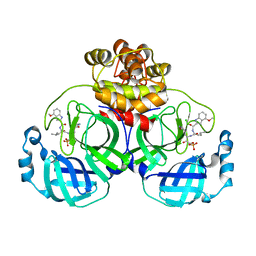 | | Crystal structure of SARS coronavirus main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SARS coronavirus main protease in complex with PF07304814
To Be Published
|
|
7WNZ
 
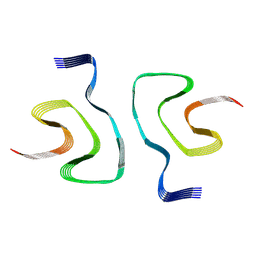 | |
3P49
 
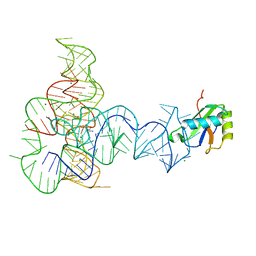 | | Crystal Structure of a Glycine Riboswitch from Fusobacterium nucleatum | | Descriptor: | GLYCINE, GLYCINE RIBOSWITCH, MAGNESIUM ION, ... | | Authors: | Butler, E.B, Wang, J, Xiong, Y, Strobel, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of cooperative ligand binding by the glycine riboswitch.
Chem.Biol., 18, 2011
|
|
6GH2
 
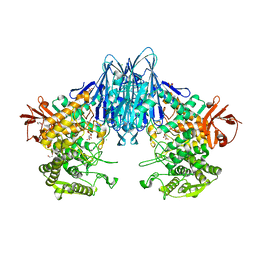 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-glc-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
