8IBI
 
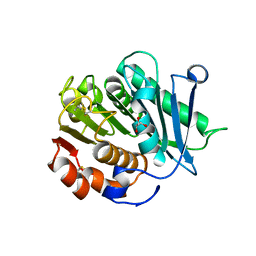 | | Inactive mutant of CtPL-H210S/F214I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
7DYS
 
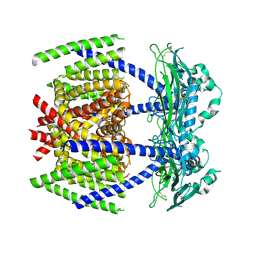 | |
3CH8
 
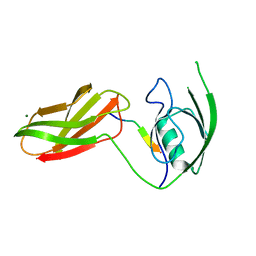 | | The crystal structure of PDZ-Fibronectin fusion protein | | Descriptor: | C-terminal octapeptide from protein ARVCF, MAGNESIUM ION, fusion protein PDZ-Fibronectin,Fibronectin | | Authors: | Makabe, K, Huang, J, Koide, A, Koide, S. | | Deposit date: | 2008-03-08 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution.
J.Mol.Biol., 392, 2009
|
|
8IBJ
 
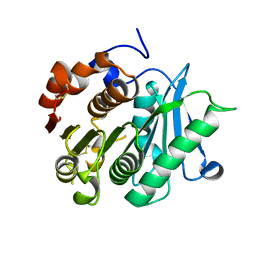 | | Inactive mutant of CtPL-H210S/F214I/N181A/F235L | | Descriptor: | PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IAN
 
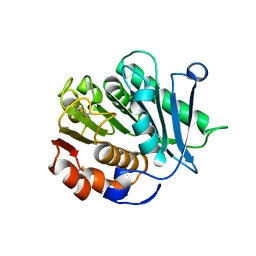 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
4Z5V
 
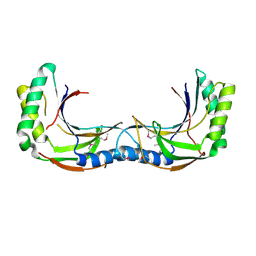 | |
4L19
 
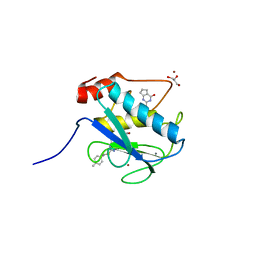 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound Q1 | | Descriptor: | 2-[(4-methylbenzyl)sulfanyl]-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, CALCIUM ION, Collagenase 3, ... | | Authors: | Minond, D, Spicer, T.P, Jiang, J, Taylor, A.B, Hart, P.J, Roush, W.R, Fields, G.B, Hodder, P.S. | | Deposit date: | 2013-06-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Characterization of selective exosite-binding inhibitors of matrix metalloproteinase 13 that prevent articular cartilage degradation in vitro.
J.Med.Chem., 57, 2014
|
|
4Z61
 
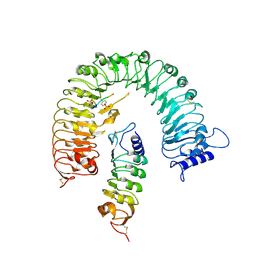 | | The plant peptide hormone receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-ILE-PTR-THR-GLN, Phytosulfokine receptor 1, ... | | Authors: | Chai, J, Wang, J. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
7DVM
 
 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
3RT2
 
 | | Crystal structure of apo-PYL10 | | Descriptor: | Abscisic acid receptor PYL10 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
4ID7
 
 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7ELF
 
 | |
3CMZ
 
 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
7W0D
 
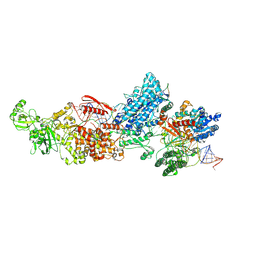 | | Dicer2-LoqsPD-dsRNA complex at mid-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0B
 
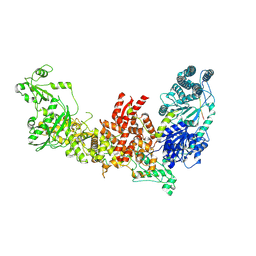 | | Dicer2-LoqsPD complex at apo status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0E
 
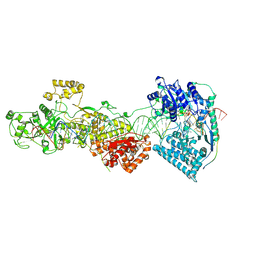 | | dmDicer2-LoqsPD-dsRNA Active-dicing status | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
5EDS
 
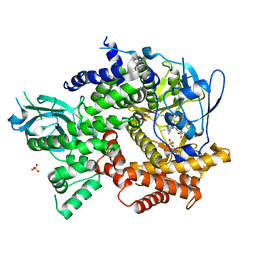 | | Crystal structure of human PI3K-gamma in complex with benzimidazole inhibitor 5 | | Descriptor: | 4-azanyl-6-[[(1~{S})-1-[6-fluoranyl-1-(3-methylsulfonylphenyl)benzimidazol-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, Optimization, and in Vivo Evaluation of Benzimidazole Derivatives AM-8508 and AM-9635 as Potent and Selective PI3K delta Inhibitors.
J.Med.Chem., 59, 2016
|
|
7W0F
 
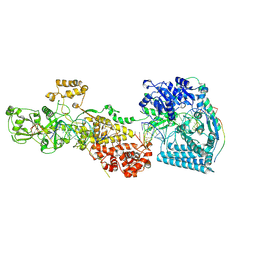 | | dmDicer2-LoqsPD-dsRNA Post-dicing status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0A
 
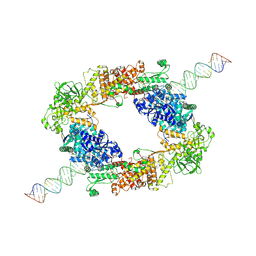 | | dmDicer2-LoqsPD-dsRNA Dimer status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0C
 
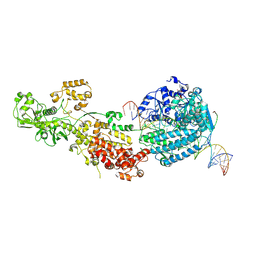 | | Dicer2-Loqs-PD-dsRNA complex at early-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
4E3R
 
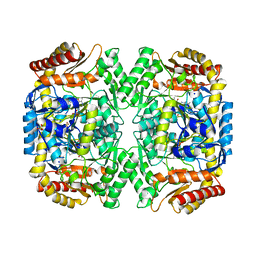 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
8IF6
 
 | |
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
3MWH
 
 | | The 1.4 Ang crystal structure of the ArsD arsenic metallochaperone provides insights into its interactions with the ArsA ATPase | | Descriptor: | Arsenical resistance operon trans-acting repressor arsD, GLYCEROL | | Authors: | Ye, J, Ajees, A.A, Yang, J, Rosen, B.P. | | Deposit date: | 2010-05-05 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The 1.4 A crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase.
Biochemistry, 49, 2010
|
|
