4QUS
 
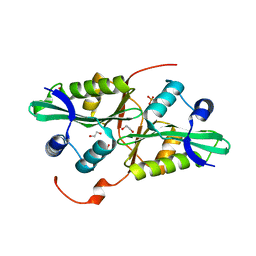 | | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase YpeA, PHOSPHATE ION | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Flores, K.J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
To be Published
|
|
4R3J
 
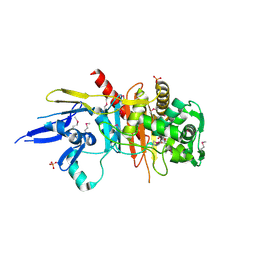 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin | | Descriptor: | (2R)-2-[(1R)-1-(acetylamino)-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin
To be Published
|
|
4R57
 
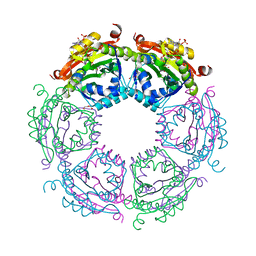 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, Spermidine n1-acetyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4R7T
 
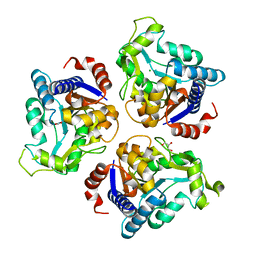 | | Crystal structure of glucosamine-6-phosphate deaminase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucosamine-6-phosphate deaminase from Vibrio cholerae
To be Published
|
|
4R3C
 
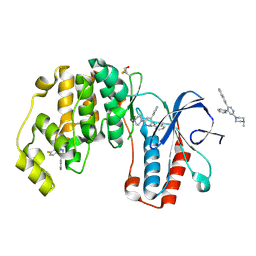 | | Crystal structure of p38 alpha MAP kinase in complex with a novel isoform selective drug candidate | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-(4-methylpiperazin-1-yl)-3-(naphthalen-2-yl)-4-(pyridin-4-yl)pyridazine, CHLORIDE ION, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting human central nervous system protein kinases: An isoform selective p38 alpha MAPK inhibitor that attenuates disease progression in Alzheimer's disease mouse models.
ACS Chem Neurosci, 6, 2015
|
|
4R86
 
 | | Crystal Structure of Aminoglycoside/Multidrug Efflux System AcrD from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-08 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of Aminoglycoside/Multidrug Efflux System AcrD from
Salmonella typhimurium
To be Published
|
|
4RCO
 
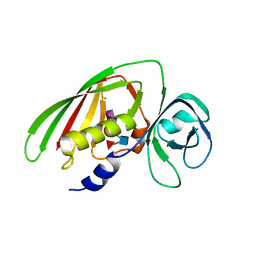 | | 1.9 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX. | | Descriptor: | CHLORIDE ION, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, Putative uncharacterized protein | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E, Halavaty, A, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX.
TO BE PUBLISHED
|
|
4QN2
 
 | | 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with NAD+ and BME-free Cys289 | | Descriptor: | ACETATE ION, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QJ1
 
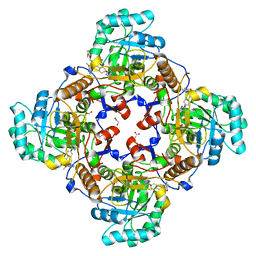 | | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109
To be Published
|
|
4QJE
 
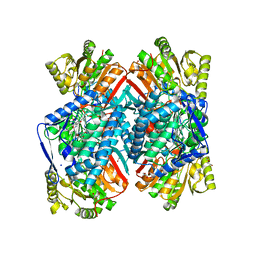 | | 1.85 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-free sulfinic acid form of Cys289 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QMK
 
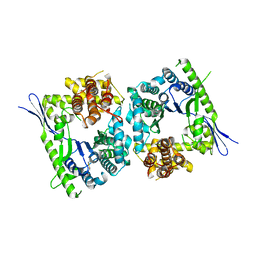 | | Crystal structure of type III effector protein ExoU (exoU) | | Descriptor: | BETA-MERCAPTOETHANOL, Type III secretion system effector protein ExoU | | Authors: | Halavaty, A.S, Tyson, G.H, Zhang, A, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Phosphatidylinositol 4,5-Bisphosphate Binding Domain Mediates Plasma Membrane Localization of ExoU and Other Patatin-like Phospholipases.
J.Biol.Chem., 290, 2015
|
|
4NMW
 
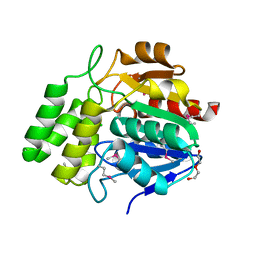 | | Crystal Structure of Carboxylesterase BioH from Salmonella enterica | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pimelyl-[acyl-carrier protein] methyl ester esterase | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Crystal Structure of Carboxylesterase BioH from Salmonella enterica
To be Published
|
|
4NOH
 
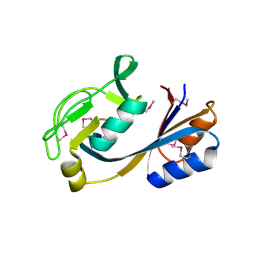 | | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Lipoprotein, putative | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4OFX
 
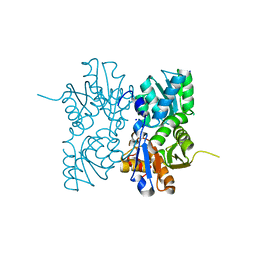 | | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii | | Descriptor: | Cystathionine beta-synthase, SODIUM ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of a Putative Cystathionine beta-Synthase from Coxiella burnetii
To be Published
|
|
4RFB
 
 | | 1.93 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with Sialyl-Lewis X. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with Sialyl-Lewis X.
TO BE PUBLISHED
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
4QVR
 
 | | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis. | | Descriptor: | Uncharacterized hypothetical protein FTT_1539c | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Ren, G, Huntley, J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.
TO BE PUBLISHED
|
|
4R7J
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1172 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni
To be Published, 2014
|
|
4PV4
 
 | | Proline aminopeptidase P II from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-14 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Proline aminopeptidase P II from Yersinia pestis
To be Published
|
|
4QWO
 
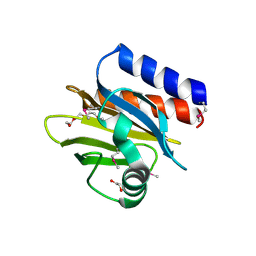 | | 1.52 Angstrom Crystal Structure of A42R Profilin-like Protein from Monkeypox Virus Zaire-96-I-16 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-06 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the Monkeypox virus profilin-like protein A42R reveals potential functional differences from cellular profilins.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
4R7R
 
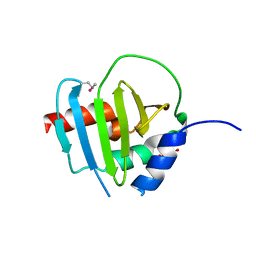 | | Crystal Structure of Putative Lipoprotein from Clostridium perfringens | | Descriptor: | GLYCEROL, Putative lipoprotein | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Crystal Structure of Putative Lipoprotein from Clostridium perfringens
To be Published
|
|
4R7Q
 
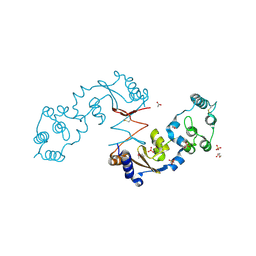 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4QVS
 
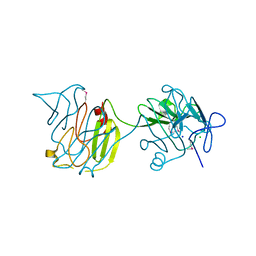 | | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405 | | Descriptor: | CHLORIDE ION, S-layer domain-containing protein, SODIUM ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405
To be Published
|
|
4R0Q
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cephalothin | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CEPHALOTHIN GROUP, Peptidoglycan glycosyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-01 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cephalothin
To be Published
|
|
4R1G
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cloxacillin | | Descriptor: | CLOXACILLIN (OPEN FORM), Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cloxacillin
To be Published
|
|
