3IZ0
 
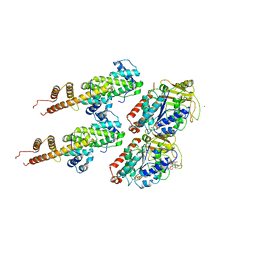 | | Human Ndc80 Bonsai Decorated Microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Ramey, V.H, Pasqualato, S, Ball, D.A, Grigorieff, N, Musacchio, A, Nogales, E. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The Ndc80 kinetochore complex forms oligomeric arrays along microtubules.
Nature, 467, 2010
|
|
3J6E
 
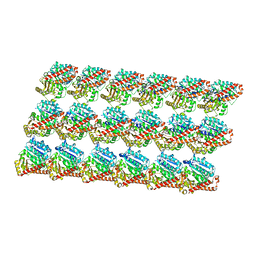 | | Energy minimized average structure of Microtubules stabilized by GmpCpp | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J6F
 
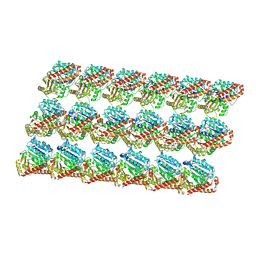 | | Minimized average structure of GDP-bound dynamic microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
2QS2
 
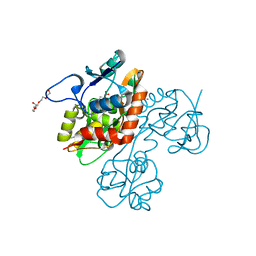 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS1
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS3
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP316 at 1.76 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-5-phenylthiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | ACET is a highly potent and specific kainate receptor antagonist: characterisation and effects on hippocampal mossy fibre function.
Neuropharmacology, 56, 2009
|
|
3J6G
 
 | | Minimized average structure of microtubules stabilized by taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Alushin, G.M, Lander, G.C, Kellogg, E.H, Zhang, R, Baker, D, Nogales, E. | | Deposit date: | 2014-02-19 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | High-Resolution Microtubule Structures Reveal the Structural Transitions in alpha beta-Tubulin upon GTP Hydrolysis.
Cell(Cambridge,Mass.), 157, 2014
|
|
6E7C
 
 | | 14-pf 3-start GMPCPP-human alpha1B/beta2B microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ti, S.C, Alushin, G.M, Kapoor, T.M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Human beta-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability.
Dev. Cell, 47, 2018
|
|
6E7B
 
 | | 13-pf 3-start GMPCPP-human alpha1B/beta3 microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ti, S.C, Alushin, G.M, Kapoor, T.M. | | Deposit date: | 2018-07-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Human beta-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability.
Dev. Cell, 47, 2018
|
|
9DVA
 
 | |
6BNW
 
 | |
7R8V
 
 | | Cryo-EM structure of the ADP state actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Espinosa de los Reyes, S, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-27 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB9
 
 | | Cryo-EM structure of the rigor state Jordan myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB8
 
 | | cryo-EM structure of the ADP state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7R91
 
 | | cryo-EM structure of the rigor state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
6BNQ
 
 | |
6UPV
 
 | | Alpha-E-catenin ABD-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Alushin, G.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism for direct actin force-sensing by alpha-catenin.
Elife, 9, 2020
|
|
6UPW
 
 | | Metavinculin ABD-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Alushin, G.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for direct actin force-sensing by alpha-catenin.
Elife, 9, 2020
|
|
7SX8
 
 | | T-Plastin-F-actin complex, parallel bundled state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural mechanism for bidirectional actin cross-linking by T-plastin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SXA
 
 | | T-Plastin-F-actin complex, pre-bundling intermediate state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (6.87 Å) | | Cite: | Structural mechanism for bidirectional actin cross-linking by T-plastin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SX9
 
 | | T-Plastin-F-actin complex, anti-parallel bundled state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mei, L, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism for bidirectional actin cross-linking by T-plastin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UDT
 
 | | cryo-EM structure of the rigor state wild type myosin-15-F-actin complex (symmetry expansion and re-centering) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-03-20 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7UDU
 
 | | cryo-EM structure of the ADP state wild type myosin-15-F-actin complex (symmetry expansion and re-centering) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-03-20 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
8D14
 
 | | Helical ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
