4RP5
 
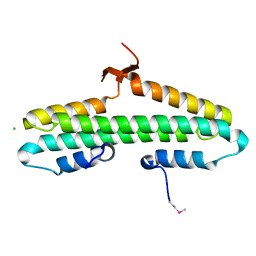 | |
4QNV
 
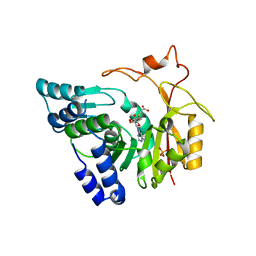 | | Crystal structure of Cx-SAM bound CmoB from E. coli in P6122 | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | 著者 | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-06-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
4RS3
 
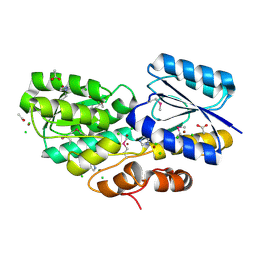 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | 分子名称: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-11-06 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4RSM
 
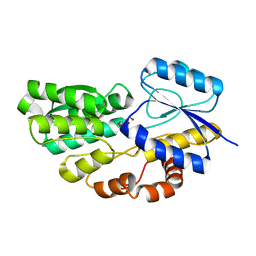 | | Crystal structure of carbohydrate transporter msmeg_3599 from mycobacterium smegmatis str. mc2 155, target efi-510970, in complex with d-threitol | | 分子名称: | D-Threitol, Periplasmic binding protein/LacI transcriptional regulator | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-11-08 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4RK9
 
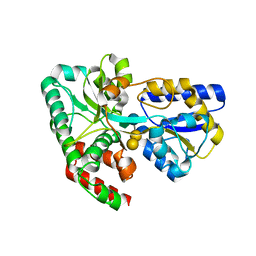 | | CRYSTAL STRUCTURE OF SUGAR TRANSPORTER BL01359 FROM Bacillus licheniformis, TARGET EFI-510856, IN COMPLEX WITH STACHYOSE | | 分子名称: | Carbohydrate ABC transporter substrate-binding protein MsmE, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-[beta-D-fructofuranose-(2-1)]alpha-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-10-12 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF SUGAR TRANSPORTER BL01359 FROM Bacillus licheniformis, TARGET EFI-510856
To be Published
|
|
4RY8
 
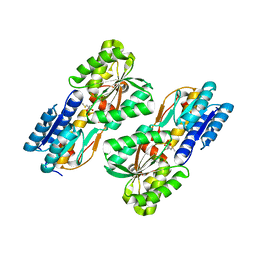 | | Crystal structure of 5-methylthioribose transporter solute binding protein TLET_1677 from Thermotoga lettingae TMO TARGET EFI-511109 in complex with 5-methylthioribose | | 分子名称: | 5-S-methyl-5-thio-alpha-D-ribofuranose, Periplasmic binding protein | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-12-13 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of 5-methylthioribose binding protein TLET_1677 from Thermotoga lettingae TARGET EFI-511109
To be Published
|
|
4R6K
 
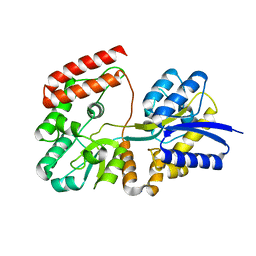 | | Crystal structure of ABC transporter substrate-binding protein YesO from Bacillus subtilis, TARGET EFI-510761, an open conformation | | 分子名称: | SODIUM ION, SOLUTE-BINDING PROTEIN | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-25 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of transporter Yeso from Bacillus subtilis, Target Efi-510761
To be Published
|
|
4R7W
 
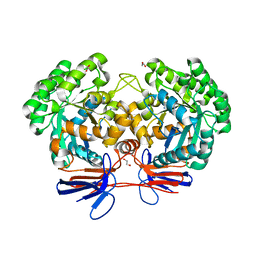 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine | | 分子名称: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, 1,2-ETHANEDIOL, Cytosine deaminase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | 登録日 | 2014-08-28 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with phosphonocytosine
To be Published
|
|
4RDW
 
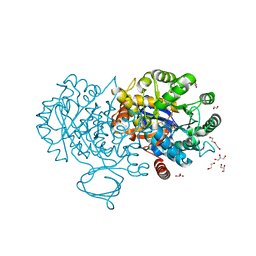 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2014-09-19 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.591 Å) | | 主引用文献 | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid
To be Published
|
|
4RXU
 
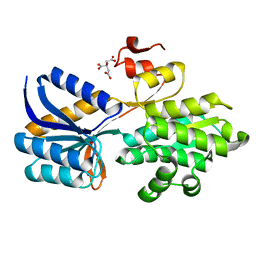 | | Crystal structure of carbohydrate transporter solute binding protein CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158, in complex with D-glucose | | 分子名称: | CITRIC ACID, Periplasmic sugar-binding protein, beta-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-12-11 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of sugar transporter CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158
To be Published
|
|
4RY1
 
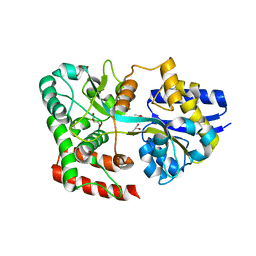 | | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum SCRI1043, Target EFI-510858 | | 分子名称: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-12-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum, Target EFI-510858
To be Published
|
|
4RWH
 
 | | Crystal structure of T cell costimulatory ligand B7-1 (CD80) | | 分子名称: | T-lymphocyte activation antigen CD80 | | 著者 | Fedorov, A.A, Fedorov, E.V, Samanta, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | 登録日 | 2014-12-04 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Crystal structure of T cell costimulatory ligand B7-1 (CD80)
To be Published
|
|
4RK1
 
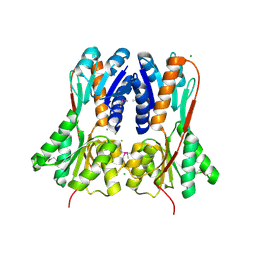 | | Crystal structure of LacI family transcriptional regulator from Enterococcus faecium, Target EFI-512930, with bound ribose | | 分子名称: | CHLORIDE ION, Ribose transcriptional regulator, alpha-D-ribofuranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-10-11 | | 公開日 | 2014-10-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of LacI Transcriptional Regulator Rbsr from Enterococcus faecium, Target EFI-512930
To be Published
|
|
4RP4
 
 | |
4RU1
 
 | | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus 11B, TARGET EFI-510965, in complex with myo-inositol | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CITRIC ACID, Monosaccharide ABC transporter substrate-binding protein, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-11-17 | | 公開日 | 2014-12-17 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
To be Published
|
|
3I0T
 
 | | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans | | 分子名称: | BH3703 protein, SULFATE ION | | 著者 | Ramagopal, U.A, Toro, R, Wasserman, S, Burley, S.K, Almo, S.C. | | 登録日 | 2009-06-25 | | 公開日 | 2009-07-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans
To be published
|
|
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | 分子名称: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | 著者 | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-01 | | 公開日 | 2009-07-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
3I6E
 
 | | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM Ruegeria pomeroyi. | | 分子名称: | MAGNESIUM ION, Muconate cycloisomerase I, SODIUM ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-07 | | 公開日 | 2009-07-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of muconate lactonizing enzyme from Ruegeria pomeroyi.
To be Published
|
|
3IJI
 
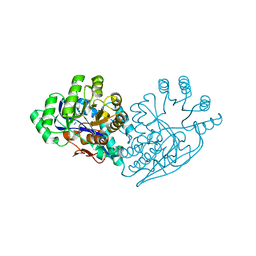 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; nonproductive substrate binding. | | 分子名称: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-08-04 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IK4
 
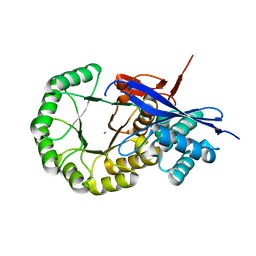 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | 分子名称: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | 著者 | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-08-05 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3I6V
 
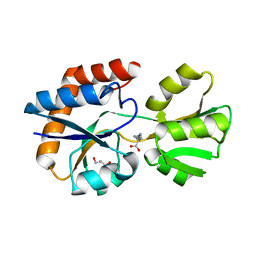 | | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine | | 分子名称: | GLYCEROL, LYSINE, SODIUM ION, ... | | 著者 | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-07 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine
To be Published
|
|
3IA1
 
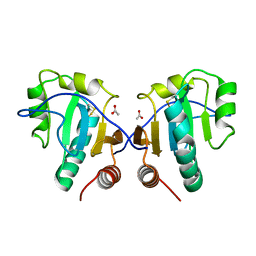 | | Crystal structure of thio-disulfide isomerase from Thermus thermophilus | | 分子名称: | ACETATE ION, Thio-disulfide isomerase/thioredoxin | | 著者 | Patskovsky, Y, Toro, R, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-13 | | 公開日 | 2009-07-21 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of thio-disulfide isomerase from Thermus thermophilus
To be Published
|
|
3IB6
 
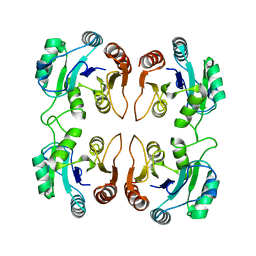 | | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b | | 分子名称: | Uncharacterized protein | | 著者 | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-15 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b
To be Published
|
|
3ID7
 
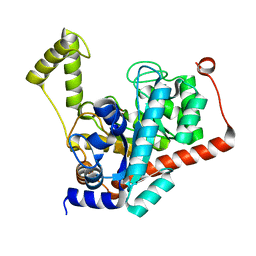 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | 分子名称: | CHLORIDE ION, Dipeptidase, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-07-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3IJ6
 
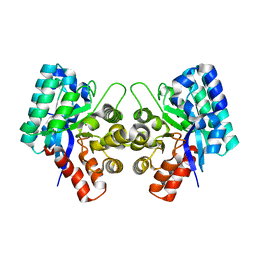 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidophilus | | 分子名称: | SODIUM ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE, ZINC ION | | 著者 | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-08-03 | | 公開日 | 2009-08-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED METAL-DEPENDENT HYDROLASE FROM Lactobacillus acidopphilus
To be Published
|
|
