2GYI
 
 | | DESIGN, SYNTHESIS, AND CHARACTERIZATION OF A POTENT XYLOSE ISOMERASE INHIBITOR, D-THREONOHYDROXAMIC ACID, AND HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHIC STRUCTURE OF THE ENZYME-INHIBITOR COMPLEX | | Descriptor: | 2,3,4,N-TETRAHYDROXY-BUTYRIMIDIC ACID, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-09-01 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of a Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, and High-Resolution X-Ray Crystallographic Structure of the Enzyme-Inhibitor Complex
Biochemistry, 34, 1995
|
|
3U3H
 
 | | X-Ray Crystallographic Analysis of D-Xylose Isomerase-Catalyzed Isomerization of (R)-Glyceraldehyde | | Descriptor: | (2R)-propane-1,1,2,3-tetrol, (4R)-2-METHYLPENTANE-2,4-DIOL, FORMIC ACID, ... | | Authors: | Allen, K.N, Silvaggi, N.R, Toteva, M.M, Richard, J.P. | | Deposit date: | 2011-10-05 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Binding Energy and Catalysis by d-Xylose Isomerase: Kinetic, Product, and X-ray Crystallographic Analysis of Enzyme-Catalyzed Isomerization of (R)-Glyceraldehyde.
Biochemistry, 50, 2011
|
|
1XYM
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | D-glucose, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
1XYL
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
5V8U
 
 | |
5V8R
 
 | |
5V8P
 
 | |
3R4C
 
 | | Divergence of Structure and Function Among Phosphatases of the Haloalkanoate (HAD) Enzyme Superfamily: Analysis of BT1666 from Bacteroides thetaiotaomicron | | Descriptor: | Hydrolase, haloacid dehalogenase-like hydrolase, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lu, Z, Dunaway-Mariano, D. | | Deposit date: | 2011-03-17 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The X-ray crystallographic structure and specificity profile of HAD superfamily phosphohydrolase BT1666: Comparison of paralogous functions in B. thetaiotaomicron.
Proteins, 79, 2011
|
|
5HI0
 
 | | The Substrate Binding Mode and Chemical Basis of a Reaction Specificity Switch in Oxalate Decarboxylase | | Descriptor: | COBALT (II) ION, OXALATE ION, Oxalate decarboxylase OxdC, ... | | Authors: | Zhu, W, Easthon, L.M, Reinhardt, L.A, Tu, C, Cohen, S.E, Silverman, D.N, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Substrate Binding Mode and Molecular Basis of a Specificity Switch in Oxalate Decarboxylase.
Biochemistry, 55, 2016
|
|
8G1N
 
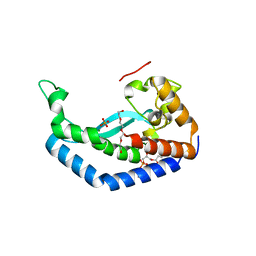 | | Structure of Campylobacter concisus PglC I57M/Q175M Variant with modeled C-terminus | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MAGNESIUM ION, N,N'-diacetylbacilliosaminyl-1-phosphate transferase, ... | | Authors: | Dodge, G.J, Ray, L.C, Das, D, Imperiali, B, Allen, K.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Co-conserved sequence motifs are predictive of substrate specificity in a family of monotopic phosphoglycosyl transferases.
Protein Sci., 32, 2023
|
|
3BGT
 
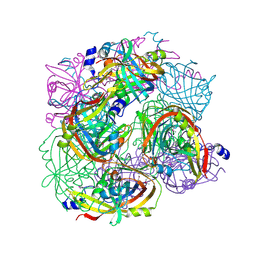 | | Structural Studies of Acetoacetate Decarboxylase | | Descriptor: | Probable acetoacetate decarboxylase | | Authors: | Ho, M, Allen, K.N. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The origin of the electrostatic perturbation in acetoacetate decarboxylase.
Nature, 459, 2009
|
|
3BH3
 
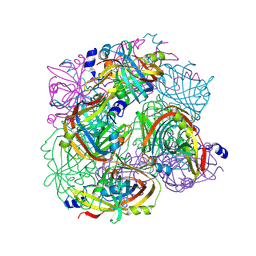 | |
3BOK
 
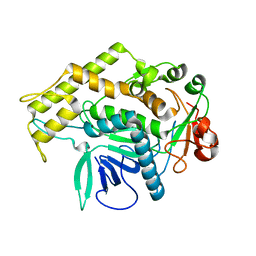 | |
3BV4
 
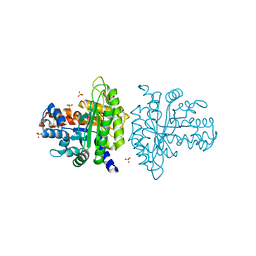 | | Crystal structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase A, SULFATE ION | | Authors: | Sherawat, M, Tolan, D.R, Allen, K.N. | | Deposit date: | 2008-01-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a rabbit muscle fructose-1,6-bisphosphate aldolase A dimer variant.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6ALD
 
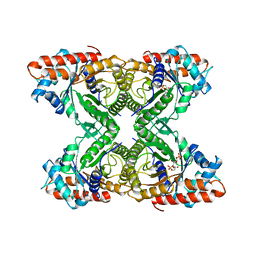 | | RABBIT MUSCLE ALDOLASE A/FRUCTOSE-1,6-BISPHOSPHATE COMPLEX | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-1,6-BIS(PHOSPHATE) ALDOLASE | | Authors: | Choi, K.H, Mazurkie, A.S, Morris, A.J, Utheza, D, Tolan, D.R, Allen, K.N. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).
Biochemistry, 38, 1999
|
|
8DB9
 
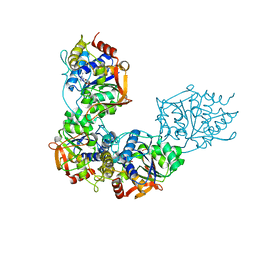 | | Adenosine/guanosine nucleoside hydrolase bound to inhibitor | | Descriptor: | 1-beta-D-ribofuranosyl-1H-1,2,4-triazole-3-carboximidamide, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8DB6
 
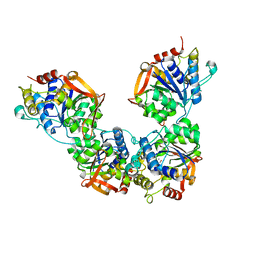 | | Adenosine/guanosine nucleoside hydrolase | | Descriptor: | CALCIUM ION, GLYCEROL, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8DB8
 
 | | Adenosine/guanosine nucleoside hydrolase bound to ImH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8DB7
 
 | | Adenosine/guanosine nucleoside hydrolase bound to a fragment inhibitor | | Descriptor: | CALCIUM ION, GLYCEROL, Inosine-uridine preferring nucleoside hydrolase family protein, ... | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
1FEZ
 
 | | THE CRYSTAL STRUCTURE OF BACILLUS CEREUS PHOSPHONOACETALDEHYDE HYDROLASE COMPLEXED WITH TUNGSTATE, A PRODUCT ANALOG | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE HYDROLASE, TUNGSTATE(VI)ION | | Authors: | Morais, M.C, Zhang, W, Baker, A.S, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2000-07-24 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily.
Biochemistry, 39, 2000
|
|
1J4E
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE COVALENTLY BOUND TO THE SUBSTRATE DIHYDROXYACETONE PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE A | | Authors: | Choi, K.H, Shi, J, Hopkins, C.E, Tolan, D.R, Allen, K.N. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
Biochemistry, 40, 2001
|
|
2ODA
 
 | | Crystal Structure of PSPTO_2114 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein PSPTO_2114, MAGNESIUM ION | | Authors: | Peisach, E, Allen, K.N, Dunaway-Mariano, D, Wang, L, Burroughs, A.M, Aravind, L. | | Deposit date: | 2006-12-22 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystallographic structure and activity analysis of a Pseudomonas-specific subfamily of the HAD enzyme superfamily evidences a novel biochemical function.
Proteins, 70, 2007
|
|
6P2D
 
 | | Structure of mouse ketohexokinase-C in complex with fructose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Michaelis-like complex of mouse ketohexokinase isoform C
ACTA CRYSTALLOGR.,SECT.D, 2024
|
|
3FM9
 
 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
