3U3H
 
 | | X-Ray Crystallographic Analysis of D-Xylose Isomerase-Catalyzed Isomerization of (R)-Glyceraldehyde | | Descriptor: | (2R)-propane-1,1,2,3-tetrol, (4R)-2-METHYLPENTANE-2,4-DIOL, FORMIC ACID, ... | | Authors: | Allen, K.N, Silvaggi, N.R, Toteva, M.M, Richard, J.P. | | Deposit date: | 2011-10-05 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Binding Energy and Catalysis by d-Xylose Isomerase: Kinetic, Product, and X-ray Crystallographic Analysis of Enzyme-Catalyzed Isomerization of (R)-Glyceraldehyde.
Biochemistry, 50, 2011
|
|
3R4C
 
 | | Divergence of Structure and Function Among Phosphatases of the Haloalkanoate (HAD) Enzyme Superfamily: Analysis of BT1666 from Bacteroides thetaiotaomicron | | Descriptor: | Hydrolase, haloacid dehalogenase-like hydrolase, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lu, Z, Dunaway-Mariano, D. | | Deposit date: | 2011-03-17 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The X-ray crystallographic structure and specificity profile of HAD superfamily phosphohydrolase BT1666: Comparison of paralogous functions in B. thetaiotaomicron.
Proteins, 79, 2011
|
|
2GYI
 
 | | DESIGN, SYNTHESIS, AND CHARACTERIZATION OF A POTENT XYLOSE ISOMERASE INHIBITOR, D-THREONOHYDROXAMIC ACID, AND HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHIC STRUCTURE OF THE ENZYME-INHIBITOR COMPLEX | | Descriptor: | 2,3,4,N-TETRAHYDROXY-BUTYRIMIDIC ACID, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-09-01 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, and Characterization of a Potent Xylose Isomerase Inhibitor, D-Threonohydroxamic Acid, and High-Resolution X-Ray Crystallographic Structure of the Enzyme-Inhibitor Complex
Biochemistry, 34, 1995
|
|
1XYM
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | D-glucose, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
1XYL
 
 | | THE ROLE OF THE DIVALENT METAL ION IN SUGAR BINDING, RING OPENING, AND ISOMERIZATION BY D-XYLOSE ISOMERASE: REPLACEMENT OF A CATALYTIC METAL BY AN AMINO-ACID | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Allen, K.N, Lavie, A, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of the divalent metal ion in sugar binding, ring opening, and isomerization by D-xylose isomerase: replacement of a catalytic metal by an amino acid.
Biochemistry, 33, 1994
|
|
5V8P
 
 | |
5V8U
 
 | |
5V8R
 
 | |
4G9B
 
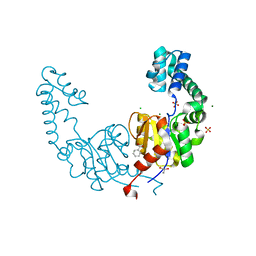 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
5WHL
 
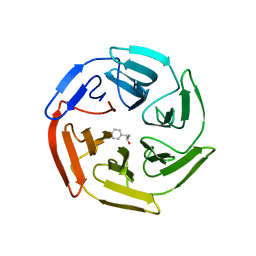 | |
5WIY
 
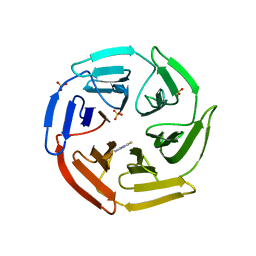 | | Kelch domain of human Keap1 bound to small molecule inhibitor fragment: 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5HI0
 
 | | The Substrate Binding Mode and Chemical Basis of a Reaction Specificity Switch in Oxalate Decarboxylase | | Descriptor: | COBALT (II) ION, OXALATE ION, Oxalate decarboxylase OxdC, ... | | Authors: | Zhu, W, Easthon, L.M, Reinhardt, L.A, Tu, C, Cohen, S.E, Silverman, D.N, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Substrate Binding Mode and Molecular Basis of a Specificity Switch in Oxalate Decarboxylase.
Biochemistry, 55, 2016
|
|
3VCA
 
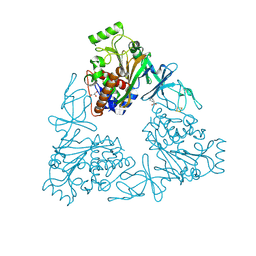 | | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman and UV-visible Spectroscopic Analysis of a Rieske-type Demethylase | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
3VDZ
 
 | | Tailoring Encodable Lanthanide-Binding Tags as MRI Contrast Agents: xq-dSE3-Ubiquitin at 2.4 Angstroms | | Descriptor: | GADOLINIUM ATOM, SULFATE ION, Ubiquitin-40S ribosomal protein S27a | | Authors: | Daughtry, K.D, Martin, L.J, Surraju, A, Imperiali, B, Allen, K.N. | | Deposit date: | 2012-01-06 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tailoring encodable lanthanide-binding tags as MRI contrast agents.
Chembiochem, 13, 2012
|
|
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
8G1N
 
 | | Structure of Campylobacter concisus PglC I57M/Q175M Variant with modeled C-terminus | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MAGNESIUM ION, N,N'-diacetylbacilliosaminyl-1-phosphate transferase, ... | | Authors: | Dodge, G.J, Ray, L.C, Das, D, Imperiali, B, Allen, K.N. | | Deposit date: | 2023-02-02 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Co-conserved sequence motifs are predictive of substrate specificity in a family of monotopic phosphoglycosyl transferases.
Protein Sci., 32, 2023
|
|
2RB5
 
 | |
2RAV
 
 | |
2RAR
 
 | |
6XCB
 
 | |
6XCC
 
 | |
3LTQ
 
 | |
2RBK
 
 | |
6XCE
 
 | |
2ODA
 
 | | Crystal Structure of PSPTO_2114 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein PSPTO_2114, MAGNESIUM ION | | Authors: | Peisach, E, Allen, K.N, Dunaway-Mariano, D, Wang, L, Burroughs, A.M, Aravind, L. | | Deposit date: | 2006-12-22 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystallographic structure and activity analysis of a Pseudomonas-specific subfamily of the HAD enzyme superfamily evidences a novel biochemical function.
Proteins, 70, 2007
|
|
