8VJX
 
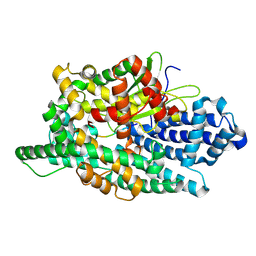 | |
8V9X
 
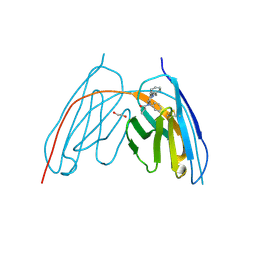 | | X-ray crystal structure of JGFN4 complex with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9Z
 
 | |
8V9Y
 
 | | X-ray crystal structure of nanobody JGFN4 | | Descriptor: | JGFN4 | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8TFD
 
 | | Crystal structure of a stem-loop DNA aptamer complexed with SARS-CoV-2 nucleocapsid protein RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DNA aptamer, Nucleoprotein | | Authors: | Esler, M, Belica, C, Shi, K, Aihara, H. | | Deposit date: | 2023-07-10 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A compact stem-loop DNA aptamer targets a uracil-binding pocket in the SARS-CoV-2 nucleocapsid RNA-binding domain.
Nucleic Acids Res., 52, 2024
|
|
5EJK
 
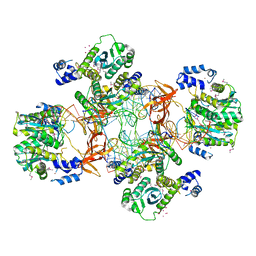 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
1Z1G
 
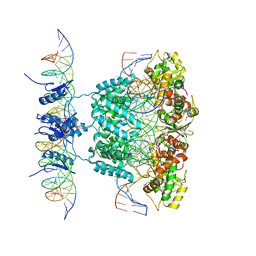 | | Crystal structure of a lambda integrase tetramer bound to a Holliday junction | | Descriptor: | 25-MER, 29-MER, 5'-D(*AP*CP*AP*GP*GP*TP*CP*AP*CP*TP*AP*TP*CP*AP*GP*TP*CP*AP*AP*AP*AP*TP*AP*CP*C)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
8SPI
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
6PAK
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Subtilisin E | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Enhancing subtilisin thermostability through a modified normalized B-factor analysis and loop-grafting strategy.
J.Biol.Chem., 294, 2019
|
|
7UU0
 
 | | Crystal structure of the BRD2-BD2 in complex with a ligand | | Descriptor: | 1,2-ETHANEDIOL, Isoform 3 of Bromodomain-containing protein 2, methyl (7S)-7-(thiophen-2-yl)-1,4-thiazepane-4-carboxylate | | Authors: | Kalra, P, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the BRD2-BD2 in complex with a ligand
To Be Published
|
|
5CQD
 
 | |
4FVA
 
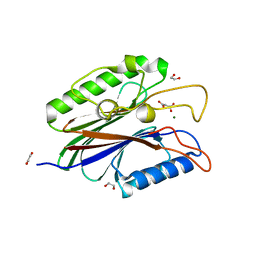 | | Crystal structure of truncated Caenorhabditis elegans TDP2 | | Descriptor: | 1,2-ETHANEDIOL, 5'-tyrosyl-DNA phosphodiesterase, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GEW
 
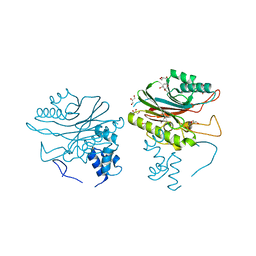 | |
5HX5
 
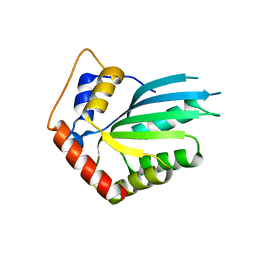 | | APOBEC3F Catalytic Domain Crystal Structure | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3F, ZINC ION | | Authors: | Shaban, N.M, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 1.92 Angstrom Zinc-Free APOBEC3F Catalytic Domain Crystal Structure.
J.Mol.Biol., 428, 2016
|
|
5HX4
 
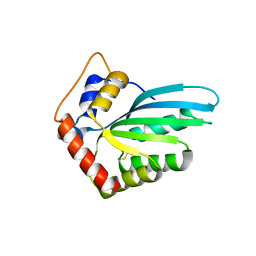 | |
6O44
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-02-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
6P7A
 
 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | Descriptor: | CADMIUM ION, Holliday junction resolvase | | Authors: | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.081 Å) | | Cite: | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
5IRS
 
 | | crystal structure of the proteasomal Rpn13 PRU-domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Chen, X, Shi, K, Walters, K, Aihara, H. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
5CQK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, GLYCEROL, SODIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
5CQH
 
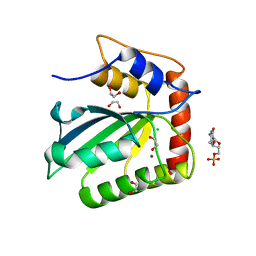 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain.
J.Biol.Chem., 290, 2015
|
|
8E5D
 
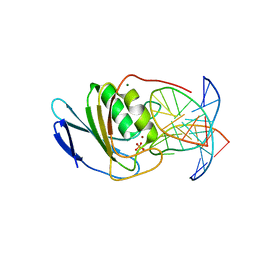 | | Crystal structure of double-stranded DNA deaminase toxin DddA in complex with DNA with the target cytosine parked in the major groove | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*CP*GP*GP*AP*CP*GP*TP*TP*GP*C)-3'), Double-stranded DNA deaminase toxin A, MAGNESIUM ION, ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis of sequence-specific cytosine deamination by double-stranded DNA deaminase toxin DddA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EJO
 
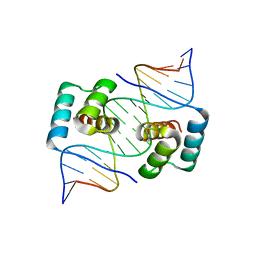 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
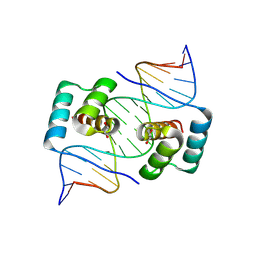 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
5CQI
 
 | |
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
